New'PPT'template - PowerPoint PPT Presentation
1 / 14
Title: New'PPT'template
1
Analysis and engineering of cellular circuitry
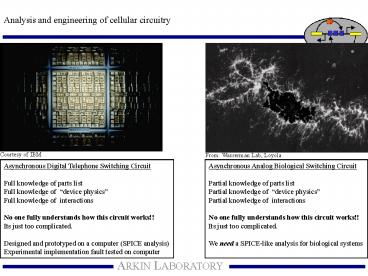
Courtesy of IBM
From Wasserman Lab, Loyola
Asynchronous Digital Telephone Switching
Circuit Full knowledge of parts list Full
knowledge of device physics Full knowledge of
interactions No one fully understands how this
circuit works!! Its just too complicated. Designe
d and prototyped on a computer (SPICE
analysis) Experimental implementation fault
tested on computer
Asynchronous Analog Biological Switching
Circuit Partial knowledge of parts list Partial
knowledge of device physics Partial knowledge
of interactions No one fully understands how
this circuit works!! Its just too
complicated. We need a SPICE-like analysis for
biological systems
2
A foundation for cell network analysis
In analogy to the steps necessary to allow
design, control and diagnosis in electronics we
must perform the following (non-sequential) tasks
Compile a list of parts that make up your
system.
Determine device physics for the parts and
their interactions.
Group subsets of parts based on function,
physics and interactions
Create databases of the above and the circuits
that are to be considered for analysis
Create quantitative simulations and analyses of
the dynamics of complex networks.
3
SPICE Simulation Program for Integrated Circuit
Evaluation
Parts database
From subcircuit database
Integrated circuit database
Automated fault diagnosis
4
BIO/SPICE Databasing, simulation and analysis
Bio/Spice A Web-Servable, Biologist-Friendly,
database, analysis and simulation interface was
developed into a true beta product. Interfaces
to ReactDB, MechDB, and ParamDB. With Kernel,
performs basic flux-balance analysis,
stochastic and deterministic kinetics, Scientific
Visualization of results. Notebook/Kernel
design optimized for distributed computing.
5
Components of Bio/Spice
BIO/SPICE
Remote Kernel
Remote Kernel
Remote Kernel
Local
Internet
Local Kernel
Center Tool Repository
Internet
Local Data
ReactDB
MechDB
ParamDB
Center Data
6
Kernel overview
- Master Simulation Controller
- Dynamically selects object level update
procedures - Schedules object updates
- Maintains global parameter and state variable
database - Manages parameter and data I/O
- Inputs
- Network definition
- User-defined new objects
- Kinetic parameters
- Model Object-Level Chemistry
- Model events internal to each object
independently - Report changes in objects output signal levels
- Update Object Parameters
- Operator combinatorial logic
- Statistical and thermodynamic parameters
- Instantaneous enzyme kinetics
- Mesoscopic Regime
- Stochastic object update procedures
- Cumulate discrete reaction events to project
signal levels
- Macroscopic Regime
- Deterministic object update procedures
- Classical integration routines to project signal
levels
Modeling procedure dynamically selected by master
controller based on current concentrations and
other model parameters
7
Bio/Spice data structure overview
World
Compartment
Pathway
Reactions
Species
Mechanisms
8
Bio/Spice Simulation Objects
Bio-Object1
Bio-Object2
Bio-Objects accept inputs in two stages 1)
resolution 2) parameterization.
Resolution determines if the output from
Bio-Object 1 is of a valid type. If it is, and
there are more than one input of that type, then
the user is prompted for which input they
mean. Parameterization determines how the
equations of both Bio-Object1 and Bio-Object2 are
affected. The user is prompted to enter the
physical data (e.g., kinetic constants) for the
interaction or to look them up in a database
9
Example of linking
User is modeling a competitive inhibition
reaction. Thus, the user creates a Reaction
Bio-Object, and four Species Bio-Objects
S
E ES EP EI
I
v kcat Etot KIS/(KSKIKSIKIS)
Linking of S to NCI-1 results in a prompt for
whether or not the species object named M1
plays the role of the internal variable, E, S, I
or P. If user chooses S, then the program
prompts for KS. If user chooses E then the
object prompts to user to enter or select all
objects in the model that have an M1 molecule
or complex within them. Currently, complex
equilibria are resolved using mass action
kinetics.
10
Using Bio/Spice to analyze phage ? Starting your
notebook
11
Using Bio/Spice to analyze phage ? Adding
infrastructure
12
Using Bio/Spice to analyze phage ? Adding the
parts
13
Using Bio/Spice to analyze phage ? Linking parts
together
14
Using Bio/Spice to analyze phage ? Entering data