Early bioenergetics: - PowerPoint PPT Presentation
1 / 18
Title:
Early bioenergetics:
Description:
Conversion of pyruvate to Acetyl-CoA (acetyl-coenzyme A). Produces CO2 ... Condenses Acetyl-CoA to yield citrate (gives the cycle its name). - Recovers CoASH. ... – PowerPoint PPT presentation
Number of Views:79
Avg rating:3.0/5.0
Title: Early bioenergetics:
1
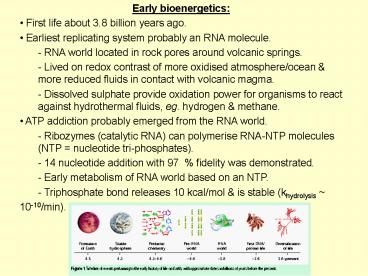
- Early bioenergetics
- First life about 3.8 billion years ago.
- Earliest replicating system probably an RNA
molecule. - - RNA world located in rock pores around volcanic
springs. - Lived on redox contrast of more oxidised
atmosphere/ocean more reduced fluids in contact
with volcanic magma. - Dissolved sulphate provide oxidation power for
organisms to react against hydrothermal fluids,
eg. hydrogen methane. - ATP addiction probably emerged from the RNA
world. - Ribozymes (catalytic RNA) can polymerise RNA-NTP
molecules (NTP nucleotide tri-phosphates). - 14 nucleotide addition with 97 fidelity was
demonstrated. - Early metabolism of RNA world based on an NTP.
- Triphosphate bond releases 10 kcal/mol is
stable (khydrolysis - 10-10/min).
2
- Cellular respiration
- Glycolysis
- - Breaks down glucose to pyruvic acid.
- Three main pathways for pyruvic acid
- - Aerobic oxidation and citric acid cycle
- Breaks down pyruvic acid to produce NADH.
- - Anaerobic alcoholic fermentation (eg. yeast).
- Break down of pyruvic acid to produce ethanol.
- - Anaerobic homolactic fermentation.
- Break down of pyruvic acid to lactate.
3
- Glycolysis
- With a few exceptions, living things all
metabolise glucose with - identical pathways.
- - In animals glucose arises in the blood from the
break down of of polysaccharides or
noncarbohydrate sources. - Glucose enters the cell's cytosol via a specific
carrier. - The enzymes of glycolysis are located in the
cytosol.
4
- Basic overview of glycolysis
- Glycolysis converts glucose to two C3 units
(pyruvate) - Ten enzymes catalyse the reactions.
- First stage (5 reactions) in which glucose
phosphorylated to yield two molecules of a
triose glyceraldehyde 3-phosphate. - Two molecules of ATP consumed.
- - Second stage (5 reactions) converts this triose
to pyruvate. - Four molecules of ATP produced.
- - Net result is to produce two ATP and two NADH.
5
- Recycling NAD
- NAD must be recycled (used as an oxidising
agent in glycolysis) - - Aerobic oxidation.
- NADH acts as an electron donor to the
respiratory chain. - - Homolactic fermentation.
- NADH used to reduce pyruvate to lactic acid.
- - Alcoholic fermentation.
- Pyruvate decarboxylated to produce
acetaldehyde, and NADH reduces this to ethanol.
6
- Citric acid cycle
- First step
- Conversion of pyruvate to Acetyl-CoA
(acetyl-coenzyme A). - Produces CO2 and one NADH (from NAD).
- Consumes CoASH (the coenzyme)
- Citrate synthasis
- - Condenses Acetyl-CoA to yield citrate (gives
the cycle its name). - - Recovers CoASH.
7
- There are eight enzymatically catalysed steps in
the cycle. - The overall reaction is
- 3 NAD FAD GDP Pi acetly-CoA
- ? 3 NADH FADH2 GTP CoA 2 CO2
- Proposed by Krebs in 1937, often called the
Kreb's cycle.
8
- Respiratory chains
- Accept electrons from an upstream donors.
- Catalyse the transfer of electrons to a terminal
electron acceptor. - Harvest the energy released to pump protons.
- - Proton gradient harvested by ATPsynthase.
9
- Mitochondria
- The workhorse'' of the cell.
- - Typically 0.7 to 1.0 mm long.
- - Outer membrane has porins allowing the free
access of small particles, metabolites etc. - Inner membrane an energy-transducing membrane.
- - About 500 mg/ml of this membrane is membrane
proteins.
10
- Mitochondrial respiratory chain
- Complex I
- - Transfers e- from NADH to quinone pool pumps
H. - Complex II
- - Transfers e- from succinate to quinone pool.
- Complex III
- - Transfers e- from quinol to cyt. c pumps H.
- Complex IV
- - Accepts e- from cyt. c, reduces O2 to H2O
pumps H. - Complex V
- - Harvests H gradient regenerates ATP
11
- Simplified bacterial respiratory chain
- The E. coli respiratory chain is simpler than
the mitochondrial. - - Lacks Complex III and cytochrome c.
- - Terminal electron acceptor takes electrons from
quinol directly. - Contains analogues to Complex I and Complex II.
- Complex IV analogue is Quinol oxidase.
- - Accepts e- from quinol, reduces O2 to H2O
pumps H. - ATP-synthase
12
- Anaerobic bacterial respiratory chain
- P. denitrificans' respiratory chain may also
donate electrons to nitrate (NO32-) or nitric
oxide (NO). - Nitrate reductase.
- Accepts electrons from quinol directly.
- Nitrate (NO32-) is reduced to nitrite (NO2-).
- Nitric oxide reductase.
- - Nitric oxide (NO) is reduced to nitrous oxide
(N2O).
Two additional soluble proteins - Nitrite
reductase catalyses NO2- ? NO. - Nitrous oxide
reductase catalyses N2O ? N2. Process called
denitrification''.
13
- Complex I of the mitochondrial respiratory chain
- Glycolysis produces 2 molecules of NADH, the
Krebs cycle another six, for each molecule of
glucose consumed. - Complex I (NADH-UQ oxidoreductase) catalyses the
2 e- transfer - from NADH to ubiquinone.
- Em,7 of the NAD/NADH couple is - 320 mV.
- Em,7 of the ubiquinone/ubiquinol couple is 60
mV. - 380 mV of energy believed used to pump 4H/2e-.
14
- Very large (about 750 kDa the large rybosome
subunit). - Contains 41 subunits.
- Bacterial analogues have only 14 subunits.
- Its redox centres (except one flavin FMN, 4Fe/S)
are eight iron-sulphur (2Fe/2S) complexes. - Cannot be studied by optical techniques.
- Requires less powerful electron spin resonance
techniques. - Only a low-resolution electron microscopy
structure. - - Shows an L-like shape.
- No clue how it pumps protons.
15
- Other methods of delivering e- to ubiquinone
- Succinate dehydrogenase (complex II)
- - Transfers electrons from succinate.
- - Part of the TCA cycle.
- ETF-ubiquinone oxidreductase (water soluble).
- - Has a surface which can dip into the membrane
which contains a ubiquinone binding site. - s,n glycerophosphate dehydrogenase.
- - Apparently similar to ETF-ubiquinone
oxidreductase. - All three are flavin proteins (FAD of FMN
prosthetic groups) with Em,7 0 mV. - Since the Em,7 of the ubiquinone/ubiquinol
couple is 60 mV, there is no net proton
translocation. - Feed electrons into the respiratory chain from
flavin linked step of fatty acid oxidation.
16
- Complex II of the mitochondrial respiratory
chain - Complex II (succinate dehydrogenase) catalyses
the 2 e- transfer from succinate to ubiquinone. - Em,7 of the Fumarate/succinate couple is 30
mV. - Em,7 of the ubiquinone/ubiquinol couple is 60
mV. - No excess energy available to pump H.
17
- Structure of Complex II
- Solved by X-ray diffraction.
- - Packs as a homo-trimer (not a functional trimer
however). - - Total molecular weight 360 kDa.
- - Four sub-units (SdhA, SdhB, SdhC, SdhD).
- Quinone binding site on cytoplasmic side.
- Redox transfer chain identified
- - Formed by FAD, 2Fe-2S, 4Fe-4S, and 3Fe-3S
clusters. - - Extends 40 Å from succinate to quinone binding
sites. - - All distances between centres lt14 Å limit for
electron transfer. - Heme b is not in the pathway.
- - Provides a handy place to store unwanted''
electrons. - - Prevents FAD from reducing O2 when quinone
deficient.
18
- Summary Lecture 3
- Glycolysis degrades glucose into two pyruvic
acid molecules. - Citric acid cycle produces three NADH and two
CO2 molecules for each pyruvate consumed. - NAD recycled for glycolysis by Complex I of
respiratory chain. - Delivers e- into the respiratory chain.
- Other pathways (eg. through complex II) for
introducing e- into the respiratory chain. - No X-ray structure exists for complex I but an
X-ray structure for complex II was solved. - More structural details are known of the
functional mechanisms of complex III, complex IV
and complex V.