From Structure to Function - PowerPoint PPT Presentation
Title: From Structure to Function
1
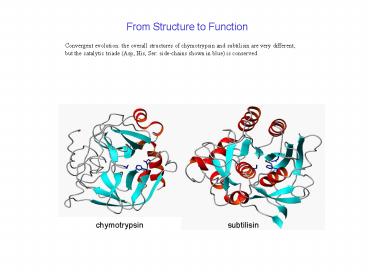
From Structure to Function
Convergent evolution the overall structures of
chymotrypsin and subtilisin are very different,
but the catalytic triade (Asp, His, Ser
side-chains shown in blue) is conserved
subtilisin
chymotrypsin
2
Enzyme mechanism
how chymotrypsin digests proteins (peptide
hydrolysis)
3
NMR spectroscopy
Determine the structure for compound C6H13NO
NMR Solvent CDCl3
1H NMR Spectra
4
2
3
1
1
1
1
13C NMR Spectra
4
NMR nuclei with spin ½ 1H 3H 3He 13C 15N 19F 29S
i 77Se 89Y 103Rh 109Ag 113Cd 119Sn 125Te 129Xe 183
W 187Os 195Pt 199Hg 207Pb
5
(No Transcript)
6
1H NMR spectroscopy of proteins
H2O
aliphatic protons
(a protease inhibitor-like protein with 60
residues)
amide and aromatic protons
7
Water in NMR spectra of proteins
NMR spectra of proteins must be recorded in H2O
(with 5-10 D2O added for the lock), because
amide protons would exchange with D2O (and their
1H NMR signal disappear). Typical protein
concentrations 0.1-1 mM Water concentration 55
M -gt dynamic range problem can be overcome by
saturating the water magnetization first,
before recording the spectrum
t
ca. 1 sec
FID free induction decay
90o pulse (a few msec)
The NMR spectrum is obtained by Fourier
transformation of the FID.
8
Because of the number of signals in the 1D NMR
spectrum, It is almost impossible to interpret.
However, two-dimensional (2D) and even 3D and 4D
spectra can be recorded which afford
the necessary resolution.
NOESY spectrum
A NOESY spectrum is recorded as a 2D data set by
recording many FIDs with systematically increment
ed t1. There are three 90o pulses in the NOESY
pulse sequence.
t2
tm
t1
In the NOESY spectrum, the diagonal peaks
correspond to the 1D NMR spectrum. Cross peaks
appear between two diagonal peaks when the
corresponding protons are close in space (lt 5
Å). If the assignment is known (i.e. which
signal belongs to which proton), this information
can be used to determine protein structures!
9
How to determine a protein structure by NMR
spectroscopy
For each NOESY cross-peak, identify the proton
pair that generates it. (This can be done with
the help of several other, more complicated two-
and multidimensional NMR spectra.) There are
several hundred NOESY cross-peaks, presenting
several hundred distance restraints (lt 5Å). Use
a computer program to find a protein structure
which fulfills all distance restraints.
LCCL domain, EMBO J. 20, 5347 (2001)
Estimating the uncertainty in an NMR structure
determination starting from several random
conformations (with bond lengths and bond angles
kept at their usual standard values), many
structures are calculated that fulfill the
experimental distance restraints. The spread
between them presents a measure of the precision
with which the structure has been determined.