Archaeal Rhodopsins - PowerPoint PPT Presentation
1 / 22
Title:
Archaeal Rhodopsins
Description:
Archaeal Rhodopsins. Four heptahelical retinal proteins identified in haloarchaea. ... methyltransferase (CheR) (reestablish an equilibrium) ... – PowerPoint PPT presentation
Number of Views:205
Avg rating:3.0/5.0
Title: Archaeal Rhodopsins
1
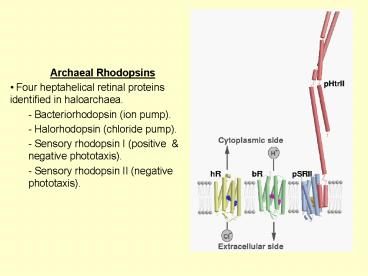
- Archaeal Rhodopsins
- Four heptahelical retinal proteins identified in
haloarchaea. - - Bacteriorhodopsin (ion pump).
- - Halorhodopsin (chloride pump).
- - Sensory rhodopsin I (positive negative
phototaxis). - - Sensory rhodopsin II (negative phototaxis).
2
- What is expressed when?
- When oxygen and respiratory substrates abundant
- - Sensory rhodopsin II is expressed.
- Absorbs near the solar maximum (500 nm).
- Initiates a negative photo-taxis signal.
- When respiratory substrates and oxygen are in
short supply - - Bacteriorhodopsin and halorhodopsin expressed.
- Energy transduction proceeds via
photosynthesis. - - Sensory rhodopsin I is expressed to optimise
light conditions. - Initiates a positive response _at_ 570 nm.
- Also has a negative response _at_ short
wavelength. - The signaling state absorbs a second photon _at_
400 nm.
3
- Phototaxis and adaption
- Bacteria typically swim in a given direction for
5 to 50 sec. - - Spontaneously tumble and change swimming
direction. - If improved conditions are found
- - Tumbling is suppressed.
- - Swim further in this direction.
- If disfavourable conditions are found
- - Tumbling is increased.
- - Looks for a new direction to swim.
- In parallel a feed-back mechanism operates which
reinstates the normal tumbling frequency. - - Called adaption.
- Combined effect is that bacteria swim up a
gradient towards favourable conditions.
4
- Signal propagation
- Structural changes in SRII conveyed to HtrII.
- A conformational change in HtrII activates the
coupling protein (CheW) a histidine kinase
(CheA). - CheA phosphorylates CheY.
- - Phosphorylated CheY a switch factor on the
Flagellar motor. - - Tumbling of bacteria suppressed or increased.
- Competitive Adaption' controlled by a
methylesterase (CheB) a - methyltransferase (CheR) (reestablish an
equilibrium). - A negative phototaxis response initiated by
SRII. - A slightly more complex dual response initiated
by SRI.
5
- Structure of sensory rhodopsin II
- Sensory rhodopsin II from Natronbacterium
pharaonis more stable than from Halobacterium
salinarium. - - Lipidic cubic phase crystals diffract to 2.1 Å
. - Fold extremely similar to bacteriorhdopsin.
- - Only helices A and B have any significant
changes relative to bacteriorhodopsin and
halorhodopsin.
6
- Spectral tuning of sensory rhodopsin II
- Sensory rhodopsin has a maximum absorption at
500 nm. - - Compare with bacteriorhodopsin (570 nm).
- Spectral tuning tailored to suit function.
- - Negative phototaxis response at solar maximum.
- Waters similar but Arg72 oriented towards
extracellular side. - - Alters the Schiff base, counter-ion
interactions. - A number of polar residues within the binding
pocket alter the polarity of the retinal
environment. - - Thr204 (Ala in bR) Val108 (Met in bR) Gly130
(Ser in bR). - - Retinal less curved in sensory rhodopsin II.
7
- K-state of sensory rhodopsin II
- As with bR, illuminate at low temperature (100
K) and determine the difference Fourier map. - - Negative density seen on Wat402.
- - More difference density peaks seen along the
retinal. - - Retinal less constrained within its pocket.
- - Also observed using FTIR spectroscopy.
8
- Why make the binding pocket less constrained
- Sensory rhodopsin II is a sensory receptor
- Requires long-lived signaling states.
- Bacteriorhodopsin requires high turnover (short
photocycle). - The signaling state of sensory rhodopsin II is
the M-state (with deprotonated retinal). - Movements of Lys205's carbonyl oxygen (Lys216 in
bR) is suppressed in sensory rhodopsin II. - Does not create sites for waters to order.
- Asp96 is absent in sensory rhodopsin II.
- Extends the lifetime of the deprotonated state.
9
- Crystal structure of complex to 2 Å
- Co-crystallised sensory rhodopsin II and a two
transmembrane helices segment (residues 1 to 114)
of the transducer. - - Packs as homo-dimer of hetro-dimers.
- Inter-helical contacts primarily hydrophobic.
- - No H-bonds between TM1 TM2 (only van der
Waals contacts) of the transducer dimer. - - TM2 of the transducer packs closely up to
helices F G of sensory rhodopsin II. - - Closer to helix G (4.06 Å average van der Waals
contacts) than helix F (4.22 Å average van der
Waals contacts).
10
- H-bonds to TM2 of HtrII
- Y199 of sensory rhodopsin II identified as
forming a H-bond to N74 of the transducer. - F-G loop (extracellular side) has three H-bonds
T189 G43 S62. - Proposed to provide Anchor points''.
- T199 very close to Pro175 (Pro185 in
bacteriorhodopsin), which provides a hinge''
about which helix F flexes. - Residues 82 to 114 not visible.
- - Cannot see if a charged patch'' interacts
with a negative patch'' on HtrII in this
region.
11
- Signaling state of sensory rhodopsin II
- A positively charged patch unique to sensory
rhodopsin II identified - from the ground state structure.
- Proposed to interact with negatively charged
residues in the transducer. - Not visible in the complex structure.
- A model for the signaling state can be built by
analogy with the bR-triple mutant structural
model of the M-intermediate. - - Show the largest movements overlap with the
charged patch.
SRII resting state
SRII signalling state model
12
- Functional Mechanism?
- Light activation of Sensory rhodopsin II
isomerises the retinal. - - Retinal pushes up upon a conserved Trp171.
- - Helix F swings out.
- - Movement is hinged near a conserved Pro175.
- Outwards tilting of cytoplasmic half of helix
proposed to - - Collide tangentially with TM2 \ induce its
rotation. - - TM2 anchored to helix G axis of rotation
between two helices. - - Rotation may unwind the cytoplasmic helices of
HtrII. - - May be the trigger which shifts the equilibrium
from inactive to active state, recognised by the
CheA.
13
- Summary
- Four rhodopsins found in halophilic archaeal
bacteria. - Display a conserved structural mechanism of
functionality. - Bacteriorhodopsin is the best understood.
- - Water molecules play a key role.
- - Steric clashes with the retinal play a key
role. - Sensory rhodopsin II (as with bacteriorhodopsin)
becoming a prototype - system of study.
- Homologues identified in eubacteria and other
prokaryotes.
14
- Structural mechanism of the the Ca2-ATPase
- Ca2 activates muscle contraction when enters
the cytoplasm. - To be effective Ca2 must be pumped out'' by
something. - Ca2-ATPase performs this task.
- - Constitutes 90 of protein within the
membrane. - P-type ATPase (include Na/K-ATPase
H/K-ATPase). - So-called since an aspartate residue is
autophosphorylated. - Two Ca2 transported per ATP consumed ( two or
three H counter transported)
8 Å Electron microscopy map of the Ca2-ATPase
15
- Ca2 ATPase crystal packing
- Crystals grown by dialysis in a mixture'' of
purified protein phospholipid
(phosphatidylcholine). - Type I crystal packing.
- Diffract to 2.6 Å.
- Distance between adjacent layers is 146 Å
- Could estimate membrane boundary from
distribution of water molecules.
16
- Protein Arrangement
- One transmembrane domain
- - Ten a-helices contain two Ca2 binding sites.
- P-domain.
- - Contains the phosphorylation residue (Asp351)
- N-domain.
- - Contains the nucleotide (ATP) binding domain.
- A-domain.
- - Act as an anchor for domain N.
17
- Ca2 Binding Site
- Two Ca2 binding sites identified
- Site I between M5 M6
- Coordinated by six oxygens side chains of a Thr,
Asn, Asp, Glu of M6, a water a Glu of M8. - Site II formed almost on'' helix M4.
- - Coordinated by six oxygens three backbone
carbonyl oxygens of M4 a Glu of M4 an Asn,
Asp of M6. - Helix M4 is unwound'' at this position (M6
also disrupted). - - Realises efficient coordination geometry rows
of oxygen atoms guide Ca2 to the binding site
remove solvating waters.
18
- ATP Binding Site
- Identified by soaking TNP-AMP into crystals.
- - Located in domain N.
- - More than 25 Å from the phosphorylation site
(Asp351). - Conformational changes must occur!
ATP site
D351
19
- Structural Changes
- Crystals grown in presence of thapsigargin (a
potent inhibitor) absence of Ca2 - - Known to lock the protein in a tight
conformation. - Data collected to 3 Å resolution.
- Molecular replacement (by moving domains
blockwise within a low-resolution EM structure)
used to solve the structure.
With Ca2
Without Ca2
20
- What changes occur?
- N domain inclines nearly 90o with respect to the
membrane. - - Equates to a 50 Å movement of the top part of
N. - P domain inclines about 30o relative to the
membrane. - - N domain inclines a further 50o relative to the
P-domain. - - P N domains virtually unchanged (rmsd 0.63
0.75 Å). - A domain rotates 110o horizontally.
- - A few hinge-residues _at_ P-A domain interface
change a lot.
21
- Changes within the membrane
- Ca2 binding site between M4, M5 M6.
- Changed conformation does not have Ca2 bound.
- Helices M1 to M6 tilt about 30o
- Helices M7 to M10 almost unchanged.
- The middle section of M5 straightens out.
- - Strongly affects L67 loop.
- - A key loop in the P-A domain interface
probably important in transmitting signal of
phosphorylation to Ca2 binding.
Purple with Ca2 Green without Ca2
22
- How does it pump Ca2?
- Muscles activated by Ca2 concentration going
up. - Ca2 enters the cytoplasmic channel binds
inducing a rearrangement of the P N domains ?
E1Ca2-state. - ATP binds between N P domains D351 is
phosphorylated, closing cytoplasmic channel ?
E1PCa2-state. - A slow conformational change occurs, opens the
luminal channel disrupts Ca2 binding ?
E2P-state 2 Ca2. - Water enters, hydrolyses the phosphoenzyme
reopens the cytoplasmic channel ? E2-state.