RECONSTRUCTING A - PowerPoint PPT Presentation
1 / 20
Title:
RECONSTRUCTING A
Description:
RECONSTRUCTING A UNIVERSAL TREE All living things share same common ancestor Classical view Prokaryotes Eukaryotes 1977: C. Woese 3 primordial kingdoms ... – PowerPoint PPT presentation
Number of Views:62
Avg rating:3.0/5.0
Title: RECONSTRUCTING A
1
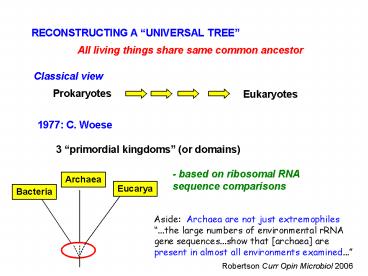
RECONSTRUCTING A UNIVERSAL TREE
All living things share same common ancestor
Classical view
Prokaryotes
Eukaryotes
1977 C. Woese
3 primordial kingdoms (or domains)
- based on ribosomal RNA sequence comparisons
Archaea
Eucarya
Bacteria
Aside Archaea are not just extremophiles
...the large numbers of environmental rRNA gene
sequences...show that archaea are present in
almost all environments examined...
Robertson Curr Opin Microbiol 2006
2
1989 Iwabe - rooting the universal tree
- if set of duplicated genes is present in all 3
lineages,
then duplication must have occurred in their
common ancestor
Fig. 5.40
- can use one gene (eg. Gene A2) as an outgroup
when comparing - the other one (Gene A1) in all 3 lineages
3
- Translational elongation factors EF-G and EF-Tu
are homologous
and both genes are present in all life forms
so ancient duplication prior to divergence of 3
superkingdoms
Fig. 5.41
Bacterial lineage diverged prior to archaeal
eukaryotic ones
4
Plant-animal-fungal trichotomy
Eucarya
Bacteria
Archaea
Fig. 5.39
Where would you place the root on this tree?
5
Maximum parsimony analysis of a tubulin
sequences
Bootstrap values above 50 are indicated above
the nodes and decay values (additional steps
needed to collapse a node) below.
all parsimony and distance-based analyses of
four large and diverse data sets support a
sister-group relationship between animals and
fungi.
Baldauf Palmer PNAS 9011558, 1993
6
ENDOSYMBIOTIC ORIGIN OF ORGANELLES
1910 - Mereschkowsky morphological similarities
between chloroplasts/mitochondria and bacteria
1960s - DNA and ribosomes discovered in
chloroplasts/mitochondria
1970 - Margulis physiological, biochemical
similarities
late 1970s - Gray, Doolittle (Halifax) -
molecular evidence for endosymbiotic origin,
from ribosomal RNA data
eg. chloroplast cyanobacterial sequences are
more similar than either is to nuclear
homologue.
7
Fig. 5.45
8
Dot divergence point of a-proteobacterial and
mitochondrial lineages
chloroplast
How do you interpret the data in this figure?
Phylogenetic tree based on SSU ribosomal RNA data
mitochondrial
Gray PNAS 86 2267, 1989
9
Protists are very diverse grouping
Tree based on ribosomal RNA data
Fig. 5.39
Certain protists (a few? lineages) lack
mitochondria
10
Did such protist lineages diverge before time of
mitochondrial endosymbiotic event?
or did they lose their mitochondria later on?
1997- 98 Mitochondrial-type genes for heat shock
proteins, etc found in nucleus of
Microsporidia, Giardia
1999 - additional sequence data places
Microsporidia within fungal clade
2003 Giardia actually has remnant mitochondria
mitosome
Nature 426 172, 2003
11
Brown Nat Rev Genet 4121, 2003
12
Evolutionary pathway for origin of eukaryotic cell
Many genes transferred to nucleus
a few retained in organelle
others lost
Alberts Fig. 14-56
13
Chimeric nature of eukaryotic nuclear genomes
Eukaryotic genomes have bacterial-type and
archaeal-type genes
Fig. 5.43
Certain genes for DNA replication/repair,
transcription/translation shared by archaea
eukaryotes (but absent in bacteria)
Possible explanations
1. Eukaryotic ancestor - archaeal, but
bacterial-type genes acquired through
horizontal transfer
- from organelles (bacterial endosymbiotic origin)
- more recent direct transfer from bacteria
2. Eukaryotic ancestor - chimeric fusion of
bacterial archaeal-type genomes
14
Model for origin of nucleus-cytosol
compartmentalization in the wake of
mitochondrial origin
Martin Koonin Nature 4441, 2006
15
HORIZONTAL GENE TRANSFER (p. 359-366)
- lateral transfer of genetic information from
one genome to another (eg. between two species)
Mechanisms
- Transformation
- - via free DNA (vector not essential)
2. Transduction - via bacteriophage or virus
3. Conjugation in bacteria - via conjugative
plasmid
Estimated that 10-18 of E.coli genome due to
LGT
eg. lactose operon (milk sugar lactose used as
carbon source in mammalian colon)
16
Detecting lateral gene transfer
1. Odd distribution patterns or unexpectedly high
similarity to homologues in distant species
2. Unusual nucleotide composition (eg. codon
usage bias, GC content)
3. Incongruent phylogenetic trees
B
A
C
A
B
C
True tree
Inferred tree
Fig. 7.22
17
Implications of lateral gene transfer?
1. Acquisition of new function
2. Replacement of native gene with captured
one
3. In bacteria, acquired genes for particular
function may be co-ordinately regulated (operon)
4. Acquisition may re-define ecological niche of
microbe
18
Web-of-life
Doolittle Uprooting the tree of life Sci.Amer.
28290, 2000
www.whoi.edu/cms/images/oceanus/2005/4/v43n2-teske
_edwards1en_8591.gif
Was early cellular life communal?
How rampant was (is) lateral gene transfer -
especially among microbes?
Highways of gene sharing among prokaryotes?
Transposable elements carry along foreign genes?
and Doolittle Science 284 2124, 1999
19
Informational genes machinery for transcription,
translation, DNA replication...
Operational genes housekeeping genes for
cellular processes (biosynthesis of amino acids,
fatty acids, nucleotides, cell envelope
proteins...)
Doolittle Cold Spring Harbor Symp. 2009
20
Prokaryotic evolution and the tree of life are
two different things
Bapteste et al. Biol. Direct 434, 2009
A molecular tree-of-life based on ribosomal RNA
sequence comparisons
Pace Mapping the Tree of Life Progress and
Prospects Microbiol.Mol. Biol. Rev. 73565, 2009