Chap. 7 Problem 1 - PowerPoint PPT Presentation
Title:
Chap. 7 Problem 1
Description:
... and therefore was the first promoter element to be identified by in vitro ... promoter-proximal control elements is linker-scanning mutagenesis ... – PowerPoint PPT presentation
Number of Views:117
Avg rating:3.0/5.0
Title: Chap. 7 Problem 1
1
Chap. 7 Problem 1
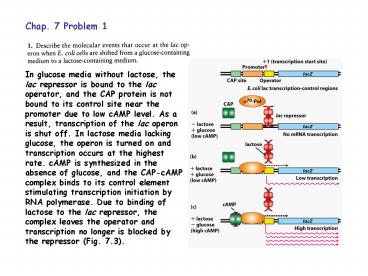
In glucose media without lactose, the lac
repressor is bound to the lac operator, and the
CAP protein is not bound to its control site near
the promoter due to low cAMP level. As a result,
transcription of the lac operon is shut off. In
lactose media lacking glucose, the operon is
turned on and transcription occurs at the highest
rate. cAMP is synthesized in the absence of
glucose, and the CAP-cAMP complex binds to its
control element stimulating transcription
initiation by RNA polymerase. Due to binding of
lactose to the lac repressor, the complex leaves
the operator and transcription no longer is
blocked by the repressor (Fig. 7.3).
2
Chap. 7 Problem 3
Part 1 Classes of RNA transcribed by RNA Pols I,
II, and III, that are important to know, are
marked with asterisks (Table 7.2). Part 2 RNA
polymerase II is very sensitive to inhibition by
the Amanita phalloides poison called ?-amanitin.
The activity of RNA Pol II, but not Pols I III
is inhibited at a 1 ?g/ml concentration.
Therefore, one can determine if a particular gene
is transcribed by RNA Pol II by determining if 1
?g/ml ?-amanitin inhibits transcription of the
gene.
3
Chap. 7 Problem 5
TATA boxes, CpG islands, and initiators all serve
as promoters, which set the transcription start
site for genes. The TATA box is found in genes
that are strongly expressed, and therefore was
the first promoter element to be identified by in
vitro transcription assays. In addition, the
transcription start site occurs at a fixed
location downstream of a TATA box (Fig. 7.14).
4
Chap. 7 Problem 6
A commonly used method to detect
promoter-proximal control elements is
linker-scanning mutagenesis (Fig. 7.21). In this
technique, a segment of random DNA is substituted
for DNA sequences across the control region.
Reporter gene assays are used to determine if the
substitutions block transcription, indicating a
control region is present.
5
Chap. 7 Problem 7
Promoter-proximal elements typically are
functional only when close to the promoter,
whereas distal enhancers often can function at
variable distances from the promoter (Fig. 7.22).
Distal enhancers sometimes can be moved to the
other side of the gene and still regulate
transcription. Both types of sequences are bound
by transcription activators that help RNA Pol II
load onto promoters.
6
Chap. 7 Problem 8
In DNase I footprinting, DNA labeled on one
strand is incubated with a transcription factor
(TF), and the complex is treated with a small
amount of DNase I, which cleaves DNA where it is
not masked by the TF (Fig. 7.23a). A control DNA
sample lacking the TF is treated under parallel
conditions. The banding patterns from the two
samples are compared by gel electrophoresis to
locate the "footprint" region where the TF has
shielded the DNA from cleavage.
In gel-shift assays (electrophoretic mobility
shift assays) (Fig. 7.24), a labeled DNA fragment
(200-300 bp) containing the binding site is
incubated with the TF and then is run on a
polyacrylamide or agarose gel. A sample of the
DNA lacking the protein is run in parallel. Bound
DNA fragments run more slowly and are shifted to
a higher position on the gel.
7
Chap. 7 Problem 9
Transcription factors have a modular structure
consisting of at least two domains.
Transcriptional activators contain a DNA binding
domain and an activation domain. Transcriptional
repressors contain a DNA binding domain and a
repression domain. Some TFs also contain a ligand
binding domain that regulates activity. Domains
typically are joined together in a single
polypeptide by flexible linker sequences that
serve as hinges and allow conformational changes
needed for activation/repression. Some examples
of transcriptional activators are shown in Fig.
7.27.
8
Chap. 7 Problem 12
The sequential assembly of Pol II transcription
pre-initiation complex is shown in Fig. 7.17. TBP
(TATA box-binding protein) binds first and
determines where transcription will initiate.
TFIIH is the last factor to bind. TFIIH has a
helicase activity that is important in melting
DNA and generating an open complex wherein the
template strand is located within the active site
of the polymerase. TFIIH also phosphorylates the
CTD of Pol II making the enzyme highly processive.
9
Chap. 7 Problem 16
UASs are comparable to promoter-proximal elements
and enhancers in higher eukaryotes (Fig. 7.22).
10
Chap. 7 Problem 22
The four main classes of DNA-binding proteins we
have discussed are the 1) helix-loop-helix
proteins (Fig. 7.28), 2) basic zipper (bZIP)
proteins (Fig. 7.29c), 3) basic helix-loop-helix
(bHLH) proteins (Fig. 7.29d), and zinc-finger
proteins (Fig. 7.29a b). A detailed description
of their structural features is presented in the
text and lecture slides. Zinc finger TFs are the
most common type of DNA binding protein encoded
by the human genome.