Project - PowerPoint PPT Presentation
Title:
Project
Description:
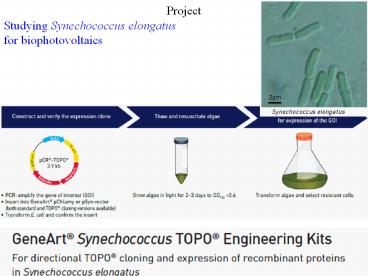
Project Studying Synechococcus elongatus for biophotovoltaics RNA Polymerase III makes ribosomal 5S and tRNA (+ some snRNA & scRNA) 100 different kinds of genes ~10% ... – PowerPoint PPT presentation
Number of Views:100
Avg rating:3.0/5.0
Title: Project
1
Project Studying Synechococcus elongatus for
biophotovoltaics
2
- How to bioengineer a novel bio-photovoltaic
system? - Obtain a sequence by PCR, then clone it into a
suitable plasmid - Were adding DNA, but want Synechococcus to make
a protein!
3
- Next Assignment
- Presentation on genome editing http//www.sciencem
ag.org/content/339/6121/768.full - Presentation on expressing a eukaryotic protein
in bacteria - Presentation on expressing a eukaryotic protein
in another eukaryote
4
- Cloning Orthologs
- 1) Identify sequence from an organism that we can
obtain - 2) Identify coding sequence
- Most sites identify Start and Stop codons
- We need 5 and 3 UTR!
- At Genbank or others can often obtain additional
flanking sequence, eg, by clicking CDS - Or, get gene name and check at TAIR, MPSS or
GRAMENE - If not, will need to obtain it based on position
- 3) Design primers to obtain entire CDS minimal
flanking sequence from suitable source!
5
- Cloning Orthologs
- Design primers to obtain entire CDS minimal
flanking sequence from suitable source! - Design nested primers that start at ATG and end
at stop codon and add CACC at 5 end - Probably will need to do this manually!
- Will break all the rules for primer design
- May get away with it if only use them on
- Amplicon
- Limited templates to bind
6
- Cloning Orthologs
- Design primers to obtain entire CDS minimal
flanking sequence from suitable source! - Design nested primers that start at ATG and end
at stop codon and add CACC at 5 end - Probably will need to do this manually!
- Will break all the rules for primer design
- May get away with it if only use them on
- Amplicon
- Limited templates to bind
- Test at http//www.idtdna.com/analyzer/application
s/oligoanalyzer/ - 5-TACTCGAAAGCAAAAGTCGTAG 62C
- 3-TTAGGCCGGGTAGCCACGC 60C
7
- Cloning Orthologs
- Design primers to obtain entire CDS minimal
flanking sequence from suitable source! - Design nested primers that start at ATG and end
at stop codon and add CACC at 5 end - Probably will need to do this manually
- Extract high MW genomic DNA from E.coli
- S. elongatus
8
- Cloning Orthologs
- Design primers to obtain entire CDS minimal
flanking sequence from suitable source! - Design nested primers that start at ATG and end
at stop codon and add CACC at 5 end - Probably will need to do this manually!
- Extract high MW genomic DNA from E.coli
- S. elongatus
- 5) Do the two PCR rxns if rxn 2 gives band of
- expected size clone it into pSyn_1/D-TOP
9
- Cloning Orthologs
- Design primers to obtain entire CDS minimal
flanking sequence from suitable source! - Design nested primers that start at ATG and end
at stop codon and add CACC at 5 end - Probably will need to do this manually!
- Extract high MW genomic DNA from E.coli
- S. elongatus
- 5) Do the two PCR rxns if rxn 2 gives band of
- expected size clone it into pSyn_1/D-TOP
- 6) Identify clones by PCR restriction digests
10
- Cloning Orthologs
- Design primers to obtain entire CDS minimal
flanking sequence from suitable source! - Design nested primers that start at ATG and end
at stop codon and add CACC at 5 end - Probably will need to do this manually!
- Extract high MW genomic DNA from E.coli
- S. elongatus
- 5) Do the two PCR rxns if rxn 2 gives band of
- expected size clone it into pSyn_1/D-TOP
- Identify clones by PCR restriction digests
- Verify by sequencing
11
- Regulating gene expression
- Goal is
- controlling
- Proteins
- How many?
- Where?
- How active?
- 8 levels (two not
- shown are mRNA
- localization prot
- degradation)
12
- Regulating gene expression
- 1) initiating
- transcription
- most important
- 50 of control
13
Microarrays Compare chromatin modification in
different tissues or treatments by
immuno-precipitating chromatin with antibodies
specific for a particular histone modification,
eg H3K4me3 then labeling precipitated DNA
controls with a different dye ChIP-chip
14
Sequencing Compare chromatin modification in
different tissues or treatments by
immuno-precipitating chromatin with antibodies
specific for a particular histone modification,
eg H3K4me3 then sequencing precipitated DNA
ChIP-seq
15
Transcription in Eukaryotes 3 RNA polymerases all
are multi-subunit complexes 5 in common 3 very
similar variable unique ones Now have Pols IV
V in plants Make siRNA
16
Transcription in Eukaryotes RNA polymerase I 13
subunits (5 3 5 unique) acts exclusively in
nucleolus to make 45S-rRNA precursor
17
- Transcription in Eukaryotes
- Pol I acts exclusively in nucleolus to make
45S-rRNA precursor - accounts for 50 of total RNA synthesis
18
- Transcription in Eukaryotes
- Pol I acts exclusively in nucleolus to make
45S-rRNA precursor - accounts for 50 of total RNA synthesis
- insensitive to ?-aminitin
19
- Transcription in Eukaryotes
- Pol I only makes 45S-rRNA precursor
- 50 of total RNA synthesis
- insensitive to ?-aminitin
- Mg2 cofactor
- Regulated _at_ initiation frequency
20
RNA polymerase I promoter is 5' to "coding
sequence" 2 elements 1) essential core includes
transcription start site
1
-100
coding sequence
UCE
core
21
RNA polymerase I promoter is 5' to "coding
sequence" 2 elements 1) essential core includes
transcription start site 2) UCE (Upstream
Control Element) at -100 stimulates
transcription 10-100x
1
-100
coding sequence
UCE
core
22
Initiation of transcription by Pol I Order of
events was determined by in vitro
reconstitution 1) UBF (upstream binding factor)
binds UCE and core element UBF is a transcription
factor DNA-binding proteins which recruit
polymerases and tell them where to begin
23
Initiation of transcription by Pol I 1) UBF binds
UCE and core element 2) SL1 (selectivity factor
1) binds UBF (not DNA) SL1 is a
coactivator proteins which bind transcription
factors and stimulate transcription
24
Initiation of transcription by Pol I 1) UBF binds
UCE and core element 2) SL1 (selectivity factor
1) binds UBF (not DNA) SL1 is a complex of 4
proteins including TBP (TATAA- binding protein)
25
Initiation of transcription by Pol I 1) UBF binds
UCE and core element 2) SL1 (selectivity factor
1) binds UBF (not DNA) 3) complex recruits Pol I
26
Initiation of transcription by Pol I 1) UBF binds
UCE and core element 2) SL1 (selectivity factor
1) binds UBF (not DNA) 3) complex recruits Pol
I 4) Pol I transcribes until it hits a
termination site
27
- Processing rRNA
- 200 bases are methylated
- Box C/D snoRNA picks sites
28
- Processing rRNA
- 200 bases are methylated
- Box C/D snoRNA picks sites
- One for each!
29
- Processing rRNA
- 200 bases are methylated
- Box C/D snoRNA picks sites
- One for each!
- 2) Another 200 are pseudo-uridylated
- Box H/ACA snoRNAs pick sites
30
- Processing rRNA
- 400 bases are methylated or pseudo-uridylated
- snoRNAs pick sites
- One for each!
- 2) 45S pre-rRNA is cut into 28S, 18S and 5.8S
products
31
- Processing rRNA
- 200 bases are methylated
- 2) 45S pre-rRNA is cut into 28S,
- 18S and 5.8S products
- 3) Ribosomes are assembled w/in nucleolus
32
RNA Polymerase III Reconstituted in vitro makes
ribosomal 5S and tRNA ( some snRNA scRNA)
33
RNA Polymerase III makes ribosomal 5S and tRNA
( some snRNA scRNA) gt100 different kinds of
genes 10 of all RNA synthesis
34
RNA Polymerase III makes ribosomal 5S and tRNA
( some snRNA scRNA) gt100 different kinds of
genes 10 of all RNA synthesis Cofactor Mn2
cf Mg2
35
RNA Polymerase III makes ribosomal 5S and tRNA
( some snRNA scRNA) gt100 different kinds of
genes 10 of all RNA synthesis Cofactor Mn2
cf Mg2 sensitive to high ?-aminitin
36
- RNA Polymerase III
- makes ribosomal 5S and tRNA (also some snRNA and
some scRNA) - Has the most subunits
37
- RNA Polymerase III
- makes ribosomal 5S and tRNA (also some snRNA and
some scRNA) - Has the most subunits
- Regulated _at_
- initiation frequency
38
Initiation of transcription by Pol III promoter
is often w/in "coding" sequence!
39
Initiation of transcription by Pol III promoter
is w/in "coding" sequence! 5S tRNA promoters
differ 5S has single C box
40
Initiation of transcription by Pol III 1) TFIIIA
binds C box in 5S 2) recruits TFIIIC
41
- Initiation of transcription by Pol III
- TFIIIA binds C box
- recruits TFIIIC
- 3) TFIIIB binds
- TBP 2 others
42
Initiation of Pol III transcription 1) TFIIIA
binds C box 2) recruits TFIIIC 3) TFIIIB
binds 4) Complex recruits Pol III
43
Initiation of Pol III transcription 1) TFIIIA
binds C box 2) recruits TFIIIC 3) TFIIIB
binds 4) Complex recruits Pol III 5) Pol III goes
until hits gt 4 T's
44
Initiation of transcription by Pol III promoter
is w/in coding sequence! 5S tRNA promoters
differ tRNA genes have A B boxes
45
Initiation of transcription by Pol III tRNA genes
have A and B boxes 1) TFIIIC binds B box
46
Initiation of transcription by Pol III tRNA genes
have A and B boxes 1) TFIIIC binds B box 2)
recruits TFIIIB
47
Initiation of transcription by Pol III 1) TFIIIC
binds box B 2) recruits TFIIIB 3) complex
recruits Pol III
48
Initiation of transcription by Pol III 1) TFIIIC
binds box B 2) recruits TFIIIB 3) complex
recruits Pol III 4) Pol III runs until hits gt 4
Ts
49
- Processing tRNA
- tRNA is trimmed
- 5 end by RNAse P
- (contains RNA)
- 3 end by RNAse Z
- Or by exonucleases
50
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- Some tRNAs are
- assembled from 2 transcripts
51
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- CCA is added to 3 end
- By tRNA nucleotidyl
- transferase (no template)
- tRNA CTP -gt tRNA-C PPitRNA-C CTP--gt tRNA-C-C
PPitRNA-C-C ATP -gt tRNA-C-C-A PPi
52
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- CCA is added to 3 end
- Many bases are modified
- Significance unclear
53
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- CCA is added to 3 end
- Many bases are modified
- No cap! -gt 5 P
- (due to 5 RNAse P cut)