RNA Splicing - PowerPoint PPT Presentation
Title:
RNA Splicing
Description:
Title: No Slide Title Author: Nigel Atkinson Last modified by: david Created Date: 10/17/2000 1:01:09 PM Document presentation format: On-screen Show (4:3) – PowerPoint PPT presentation
Number of Views:118
Avg rating:3.0/5.0
Title: RNA Splicing
1
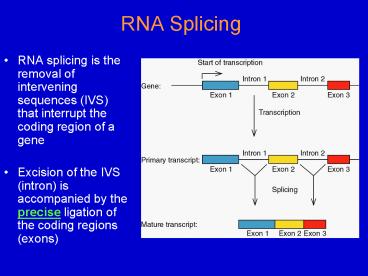
RNA Splicing
- RNA splicing is the removal of intervening
sequences (IVS) that interrupt the coding region
of a gene - Excision of the IVS (intron) is accompanied by
the precise ligation of the coding regions (exons)
2
Discovery of Split Genes (1977)
- P. Sharp and R. Roberts - 1993 Nobel Prize in
Physiology Medicine - Discovered using R-loop Analysis
- Cloned genomic DNAs of a few highly expressed
nuclear genes (e.g., hemoglobin, ovalbumin), and
certain Adenoviral genes were hybridized to RNA
fractions and visualized by EM - Loops form from RNA annealing to the template
strand and displacing coding strand of DNA
3
Genomic DNA fragment containing a Globin gene was
annealed to large heterogenous nuclear RNA
(hnRNA), which contained globin mRNA precursors.
Template strand
coding
Dotted line is RNA
DNA
Fig. 14.3a
4
template strand
Coding strand
When genomic globin gene was annealed to
cytoplasmic mRNA (which contained mature globin
mRNA) got an internal loop of single-stranded DNA
( spliced out intron).
Fig. 14.3b
5
Intron Classes Distribution
- Group I - common in organelles, nuclear rRNA
genes of lower eukaryotes, a few prokaryotes - Group II - common in organelles, also in some
prokaryotes and archaea - Nuclear mRNA (NmRNA) - ubiquitous in eucaryotes
- Nuclear tRNA- some eucaryotes
6
Relationships of the 4 Intron Classes
- Each has a distinctive structure.
- The chemistry of splicing of Groups I, II and
NmRNA is similar i.e, transesterification
reactions - The splicing pathway for Group II and nuclear
mRNA introns is similar. - Splicing of Groups I, II and possibly NmRNA
introns are RNA-catalyzed
7
Self-Splicing Introns
- Some Group I II introns self-splice in vitro in
the absence of proteins - are ribozymes. - Conserved secondary structure but not primary
sequence. - Require Mg2 to fold into a catalytically active
ribozyme. - Group I introns also require a guanosine
nucleotide in the first step.
8
Tetrahymena rRNA Group I Intron
- First self-splicing intron discovered by T.
Cechs lab in 1981 - In the 26S rRNA gene in Tetrahymena
-
- First example of a catalytic RNA!
- Nobel Prize in Chemistry to T. Cech and S.
Altman (showed that RNase P was a true
turnover riboenzyme in vivo), 1989
9
Group I splicing mechanism
GOH guanosine nucleotide, guanosine will work
because the phosphates dont participate in the
reaction. In vivo, GTP probably used.
The 3 terminal G of the intron is nearly 100
conserved.
Fig. 14.47
10
Cr.LSU intron 2ndary structure of a group I
intron
Old style drawing
Newer representation
Exon seq. in lower case and boxed
Shows how splice sites can be brought close
together by internal guide sequence.
Conserved core
5 splice site
11
RNA structures seen in Group I introns
- G ? U pairs
- Stacked helices
- Long-range base pairings (P3 and P7, also form
last) - Triple helices (3 strands) (P4-P6 junction area)
12
3-D Model of Tetrahymena rRNA Intron
Catalytic core consists of two stacked helices
domains 1. P5 P4 P6 P6a (in green) 2. P9
P7 P3 P8 (in purple) The substrate is
the P1 P10 domain (in red and black), it
contains both the 5 and 3 splice sites.
13
A two-metal ion mechanism for group I intron
splicing
(Second Step)
M. R. Stahley et al., Science 309, 1587 -1590
(2005)
14
Guanosine binding site of Group I Introns
- Mainly the G of a G-C pair in the P7 helix of
the conserved core. - Highly specific for Guanosine (Km 20 µM).
- Also binds the 3?-terminal G of the intron in
the second splicing step.
15
Splicing Factors for Self-Splicing Introns
- Some Group I and many Group II introns cant
self-splice in vitro (need protein factors) - Even self-splicing introns get help from
proteins in vivo - Based on fungal (yeast and Neurospora) mutants
deficient in splicing of mitochondrial introns
(respiratory-deficient)
16
Protein Splicing Factors for Group I (and Group
II) Introns
- 2 types
- Intron-encoded
- - promote splicing of only the intron that
encodes it - Nuclear-encoded
- - Splice organellar introns
17
Split Genes of Yeast Mitochondria
From Phil Perlman
Proteins encoded within these introns Mat
maturase (promotes splicing) Endo DNA
endonuclease, promotes intron invasion
18
- Nuclear-encoded splicing factors function by
- Promoting correct folding of the intron (cbp2)
- - CBP2 promotes folding of a cytochrome b intron
(bI5) - Stabilizing the correctly folded structure
(cyt-18) - Cyt-18 promotes splicing of a number of Mt Group
I introns in Neurospora. - Cyt-18 is also the Mt tyrosyl-tRNA synthetase,
dual- function protein. - Evolved from the tyrosyl-tRNA synthetase
by acquiring a new RNA-binding
surface.
Alan Lambowitz
19
Figure 5. Models of CYT-18/?C424-669 with Bound
RNA Substrates(A) Dimeric CYT-18/?C424-669 with
the T. thermophilus tRNATyr (orange) docked as in
the T. thermophilus TyrRS/tRNATyr cocrystal
structure (Yaremchuk et al. 2002 PDB ID 1H3E).
Subunits A (Sub. A magenta) and B (Sub. B blue)
are defined as those that bind the tRNA acceptor
and anticodon arms, respectively. Side chains at
positions that did or did not give specific
EPD-Fe-induced cleavages in the ND1 intron are
shown in space-filling representations colored
yellow and black, respectively.(B) Stereoview of
dimeric CYT-18/?C424-669 with docked ND1 intron
RNA. The model is based on optimized fit to
directed hydroxyl radical cleavage data
summarized in Figure 4B. The ND1 intron RNA
(residues 27182) is shown as a green ribbon,
with purple balls indicating phosphate-backbone
protections from full-length CYT-18 protein
(Caprara et al., 1996a), and red ribbon segments
indicating EPD-Fe cleavage sites. The C-terminal
domain of T. thermophilus TyrRS (yellow) is shown
positioned on subunit B as in the T.
thermophilus/tRNATyr cocrystal structure
(Yaremchuk et al., 2002).