RNA Splicing - PowerPoint PPT Presentation
1 / 16
Title:
RNA Splicing
Description:
An extreme example of how splicing and processing gives rise ... This experiment can be repeated for all of the other putative control regions. March 20, 2002 ... – PowerPoint PPT presentation
Number of Views:751
Avg rating:3.0/5.0
Title: RNA Splicing
1
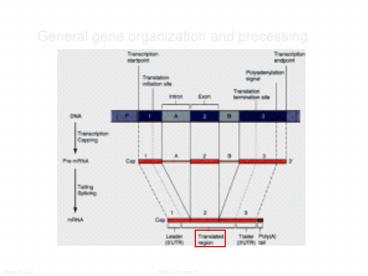
General gene organization and processing
2
Splicing can be regulated by repressors and
activators
3
An extreme example of how splicing and processing
gives rise to two completely different gene
products from the same gene - the
calcitonin/neuropeptide gene
4
Splicing enhancers can regulate alternative
splicing Bound regulatory proteins recruit key
splicing factors to weak splice sites (those
sites that can be overlooked by splicing
machinery if enhancers are not present)
5
In addition to alternative splicing, the use of
alternative promoters also generates diversity in
patterns of gene expression In this example, the
same protein is encoded but the 5 UTR region is
different. Meaning the transcript may have
different properties and be regulated in
different ways
Translation initiation site
6
There are protein-binding modules that are used
repeatedly to control the expression of different
genes Each promoter contains different numbers
and arrangements
7
An example of regulation of gene expression
A Drosophila embryo in which the expression
pattern of two genes is highlighted- yellow is
ftz and blue is even-skipped (eve) Their
expression is regulated by an elaborate network
of activators and repressors
8
The eve promoter has multiple stripe
modules They were found by cloning segments of
the promoter region into a vector that drives
expression of lacZ
9
Generation of a transgenic fly expressing the eve
stripe 2 promoterlacZ results in an embryo in
which lacZ is seen only in the area where the
normal eve strip 2 would be seen This experiment
can be repeated for all of the other putative
control regions
10
Experiments showing that strip 1 and 5 could also
be specified by a small region of the promoter
11
The eve stripe two control region has binding
sites for 4 regulators- -activators Bicoid and
Hunchback -repressors Giant and Kruppel The
spatial expression of these regulators determines
if the gene is expressed
on previous slide, look at what happens in a
Giant mutant- get expansion of eve expression!
12
The promoter region of the rat PEPCK gene
contains many binding sites for transcriptional
regulators -among them some leucine zipper
dimers and some other homo-dimers and
hetero-dimers -Many of these elements are
responsive to hormones
13
Micro-Array technology to analyze gene expression
The principle behind this is to look at
differences in gene expression when variables are
changed - eg. Yeast cells grown in the presence
of EtOH- what genes are turned on or off in
response to that change in the environment
Pool the cDNAs
14
The cDNAs will be hybridized to microarrays on
which every gene that has been cloned is present
the DNA is spotted on the microslides and each
spot corresponds to DNA from a different
gene If a particulatr gene is expressed, then
it will be present and labelled in the the cDNA
pool. It can then hybridize to the spot of the
plate corresponding to that particular gene
15
The results from such an experiment look like
this where the colour of the spot tells you
something about that gene expression.
16
The data can then be analyzed and sorted into
tables that show which genes are expressed in
response to the stimulus and which are turned
off This sort of experiment can be done with any
collection of RNAs that you want to compare-
particularly useful to compare normal to
mutant/disease state- eg. tells you what genes
are turned on in cancerous cells, may give you a
clue as to how cancer works