Chapter 7: Why mutations are important - PowerPoint PPT Presentation
1 / 36
Title:
Chapter 7: Why mutations are important
Description:
... sequence and function in mice and humans, but is ... Compare mice and man again... Gene X Gene X Promoter 1. Promoter 2. human. mouse (in brain) ... – PowerPoint PPT presentation
Number of Views:63
Avg rating:3.0/5.0
Title: Chapter 7: Why mutations are important
1
S21
Chapter 7 Why mutations are important -the stuff
of experimental genetics -the stuff of
evolution!!! The capacity to blunder slightly is
the marvel of DNA. Without this special
attribute, we would still be anaerobic bacteria
and there would be no music. Lewis Thomas,
biology watcher/philosopher
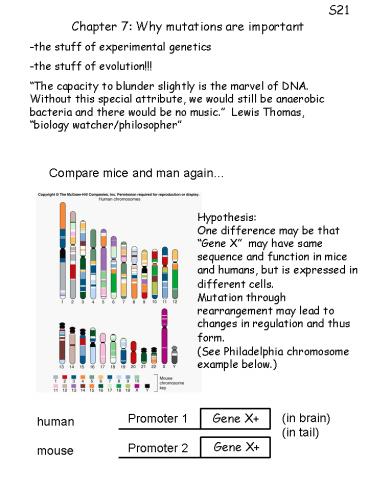
Compare mice and man again
Hypothesis One difference may be that Gene X
may have same sequence and function in mice and
humans, but is expressed in different cells.
Mutation through rearrangement may lead to
changes in regulation and thus form. (See
Philadelphia chromosome example below.)
(in brain) (in tail)
Promoter 1
Gene X
human mouse
Gene X
Promoter 2
2
S22
What is a Mutation? Base-pair change that
alters the behavior of a protein, cell,organism.
Forward and reverse mutation Forward A
to a- Reverse a- to A
Forward mutation More common than reverse
mutation
A
ORF
Many potential sites of mutation
a-
ORF
Usually one (of very few) sites to reactivate a-
ORF
Base-pair changes changes can either be silent or
disruptive. 1. Some amino acids are important to
protein function, some not. 2. Cannot usually
say a priori if a base pair difference leading to
an amino acid difference between 2 alleles is
important!
Consider immunoglobulin fold
3
Famous Mutations 1 Fragile X
S23
Causes mental retardation due to trinucleotide
expansion
FMR-1 genes in unaffected people have fewer
than 50 CGG repeats. Unstable premutation
alleles have between 50 and 200 repeats.
Disease causing alleles have gt 200 CGG
repeats.
Pedigree shows X- linkage
Nucleotide expansion inactivates FMR-1 and causes
chromosome fragility..
Normal X mutant X
4
S24
Famous Mutations 2 The Philadelphia Chromosome
and the Bcr-Abl fusion in CML
A. Reciprocal translocation between Chr9 and Chr
22
B. Chromosome fusion alters regulation of Abl
protein kinase
Normal Mutant
C. Gleevec inhibitor works!
5
S25
Kinds and mechanisms of mutation
substitution
Fig 7.2
deletion
insertion
Translocation (example Philadelphia chromosome!)
6
S26
Mutation rate Roughly 0.1-0.6 mutations/haploid
genome (1 mutation potentially affecting
phenotype in every 2-10 human gametes)
Fig 7.3
- ..dedicated researchers monitored coat color
(arising spontaneously) in progeny.. - (This is the forward rate, of course)
- Rates of mutation vary from gene to gene due to
- DNA sequence.
- size of gene.
- mutability of protein to amino acid changes.
7
S27
Luria-Delbruck Fluctuation Test
Do mutations in a gene occur as a physiological
response to stress, or do they occur at random in
time?
Test 1 Fluctuation Analysis
2 hypotheses and unique predictions of each
1. Resistance is a physiological response
1. Resistance by mutation is a physiological
response
2. Resistance by mutation arises randomly in time
Results fit with expectations if random mutation
occur at random.
8
S28
Test 2 Mutations pre-exist before stress
9
Kinds and mechanisms of mutation
S29
Fig 7.6
- Depurination- 1000/hr/cell!!
- Deamination-CgtU, causing potentially GCgtAT
mutation - (Special enzymes remove U from DNA to
limit mutation.) - X-rays, UV, oxidation
10
S30
How mutagens alter DNA Fig. 7.12
one example
11
S31
XP individuals (xeroderma pigmentosum)
10-15 proteins
12
S32
Ames Test- one easy way (?) to determine if a
chemical can mutate DNA
Q reverse of forward mutation? Another point
There are issues
13
S33
Mutations Larger scale changes
- Philadelphia chromosomeagain..
- One mechanism shown Fig 7.10
One chromosome has one hybrid gene, Other
chromosome has two normal genes and one hybrid
Homologous Recombination
- Identical sequences may align and may then
recombine.
a c g g c a t t a g g c c a t g c a t g c g c c
c c a a
t g c c g t a a t c c g g t a c g t a c g c g g g
g t t
a c g g c a t t a g g c c a t g c a t g c g c c
c c a a
t g c c g t a a t c c g g t a c g t a c g c g g g
t t t
g c g t a t c c g a t c a g g g c c
c g c a t a g g c t a g t c c c g g
RecA protein is the magical element that promotes
homologous pairing
14
S34
Next A basic genetics question You have
isolated two mutations that have same
phenotype Are two mutations in the same or
different genes??
Wt plus 3 mutants
Induce mutations (by feeding mutagen (EMS) for
generations).
Complementation tests or Recombination tests
15
S35
Are m1 and m2 in same or different gene (Fig
7.15.)
Complementation Test
Cross m1 (m1/m1) to m2 (m2/m2).
If progeny with both mutations have mutant eyes,
then mutations fail to complement, and mutations
are in same gene (most likely).
If progeny with both mutations have normal eyes,
then mutations complement, and
mutations are in different genes (most
likely).
m1/m2
m1/ m2/
Table white and cherry are two alleles of
same gene, while white and garnet are two alleles
of different genes..and so on.
ISSUE complementation tests only useful when
both mutations are recessive!
16
Complementation tests with recessive
mutations Mutations recessive to wildtype (and
m1/m1 and m2/m2 mutant, of course).
S36
Genotype Phenotype
Same Gene
1
(m1/)
2
(m2/)
3
4
in same gene
-
(m1/m2)
Different Genes
5
These are the different!
6
7
8
in different genes
(m1/, m2/)
17
Complementation tests with a dominant
mutation(s) Result is not interpretable!
S37
Same Gene
Genotype Phenotype
1
2
m1/
3
-
m2/
4
-
m1/m2
Different Gene
5
These are the same!
6
m1/
7
-
m2/
-
8
18
S38
On the nature of dominant and recessive
mutations.
Fig 7-25
Typical case mutations recessive
Less-usual case mutation1 dominant
(haploinsufficiency)
19
S39
On the nature of mutations incompletedominance
Fig 7-26
20
S40
Other explanations for dominant mutations
d. Ectopic expression making protein in a cell
its not normally made.
b. One explanation for dominant negative mutations
21
S41
Second method to determine if mutations in same
or different gene Recombination
If mutations in same gene, then closelinked,
and few wt recombinants
(less DNA between alleles where recombination can
occur)
Meiosis recombination
If mutations in different genes, then more wt
recombinants (more DNA between alleles were
recombination can occur)
Meiosis recombination
22
S42
Bacteriophage the Genetic Code
Phage and its life cycle
Simple techniques
23
S43
Key phenotypes of rII mutants
Genotype Phenotypes
Permissive host Restrictive host
Plaque morphology on strain B
24
S44
Complementation tests using phage
FIG 7-17
one step test NO RECOMBINATION INVOLVED
25
S45
Recombination tests using phage
More realistic result, for example
recombination test d1. 100 plaques
control d2. 1
plaque
(Control test for forward or reverse mutation?)
26
S46
Genetic mapping with phage
Why are there hot spots of mutation?
27
S47
All Hail the Genetic Code!!
28
S48
- Test of Genetic Code (pg 312-319)
- General Nature of the Genetic Code. Crick,
Barnett, Brenner, Watts-Tobin Nature 192, 1227,
1961. - Questions they address..and answer
- How many bases encode one amino acid.
- Stop codons (nonsense) and code degeneracy
- Genes read directionally.
Triplet Nature of Code
TCG ATG TCT GCA TAT CAA TGC ATC TTG ACT TAC Ser
Met Ser Ala Tyr Gln Cyc Ile
Leu Thr
in frame
Stop codons and Degeneracy are linked ideas..
Directionality
rIIA
rIIB
29
S49
B. Experimental Design Use of rII locus and
special mutagen-
proflavin
Massive dose of intuition.. They argue that
proflavin likely adds or deletes bases and does
not change a base.
Proflavin-made mutants resembled
deletion mutants.
Method Grow rII on strain B in presence of
proflavin (the mutagen) Then
plate to form plaques on B, and check each plaque
on K12
Permissive Restrictive
Strain B Strain K12 Plaque phenotype
Genotype
rII rIID (deletion) rII- (chemical change to
base) rII- (made with proflavin) rII-, sup1
(suppressor of rII) rIID, sup1
Wildtype Mutant tight Mutant fuzzy
leaky Mutant tight pseudo wildtype
(suppression) Mutant, no suppression
30
S50
Proflavin- they reasoned correctly!
Fig 7-12
31
S51
C. Isolating mutations, then suppressors, then,
and then
rIIB
Grow on B with proflavin. Plate on B and
identify that then dont grow on K
Phenotype
rII-1 rII-2
rII-1
(FCO)
Select rare suppressors on K12
mutation
On B
Coinfect B Recombination Plate on B
D. Separate rII-2 from rII-1
rII-1
rII-1 rII-2
rII-2
wt
E. Isolate suppressors of rII-2
On K12
rII-2
rII-2 rII-3
Select rare suppressors on K12
F. Separate by recombination
Coinfect B Recombination Plate on B
On B
rII-2 rII-3
rII-2
rII-3
Sum Make initial mutations isolate suppressors
separate by recombination isolate more
suppressors map mutations
32
S52
G. Sign convention of mutations and suppressor
mutations
- Genotype Phenotype sign
- B K12
- rII 0
- rII-1 (FC0) -
- rII-1 rII-2 0
- rII-2 -
- - rII-2 rII-3
0 - rII-3 -
Etc etc
H. Test of single and double mutants
Normal protein, Out
of frame, gibberish protein
rII
Full length normal rII protein
Short, or nonfunctional protein (translated out
of frame!)
rII-1
rII-1 rII-2
Full length, mostly normal, protein
rII-1 rII-4
Full length, mostly normal, protein
rII-1 rII-3
Abnormal protein, twice in wrong frame
33
S53
I. Codon frame interpretation
rII CAT CAT CAT CAT CAT CAT normal
protein rII-1 CAT ATC ATC ATC ATC ATC
gibberish rII-2 CAT CAT CAT CAT CCA TCA
TCA TCA TCA TCA
-1
1
rII-1 rII-2 CAT ATC ATC ATC CAT CAT CAT CAT
I. Test of number of bases per codon
rII
Full length normal protein
rII-1 rII-3
Abnormal protein, twice in wrong frame
,
Mostly normal protein, with some gibberish yet
protein still functional..
rII-1 rII-3 rII-5
,,
(They actually knew from earlier studies that
only the last 1/3rd of protein was structurally
important for plaque formation!!)
They did many more tests Conclusion Most ,,
yield functional protein Most -.-.- yield
functional protein All other combinations (,,-
,-,- etc) do not yield functional protein.
34
S54
K. Addressing an odd observation
Forbidden transitions suggests stop codons.
(An exception can prove a general rule!)
Observation Certain combinations of and - did
not yield K12 plaques
FCO()
101(-)
Phenotype Suppression Suppression Suppression N
O suppression
rIIB gene
A.
FC9(-)
51()
B.
40() 87(-)
C.
7(-) 47()
D.
Certain regions in rIIB gene dont allow both
- and -.
Interpretation
For A and B, see earlier. For C and D..
ATG ATG ATG ATG ATG ATG
in frame ATG AAT GAT GAT GAT
ATG ATG - ATG TGA TGA
TGA TGA ATG ATG ATG -
1
-1
Yes NO
Asn
-1
1
stop
35
S55
J. Degeneracy of genetic code Given 3
bases per codon 4 bases possible in DNA
(A,C,T,G) then 64 possible 3 base
combinations 20 amino acids Therefore,
either A. Each amino acid is encoded by
one codon, so 44 codons do not encode an amino
acid and presumably would be a STOP or
nonsense OR B. Each amino acid is encoded by
more than one codon, so most of the 64 possible
codons would be sense. A and B make different
predictions about distance between suppressor
mutations. If A is true, distance would be very
short(44/64 codons, or 2/3, would be nonsense
and stop translation and result in no plaque). If
B is true, distance could be much longer..(most
codons, but not all, that are out of frame are
still sense) They looked at genetic distance
between suppressors
36
S56
K. Directionality Again, they started with a
trick The rII locus contains two genes, rIIA and
rIIB, both of which are required to form plaques
on host K12. (All experiments up to now were done
with rIIB mutations only.)
rIIA
rIIB
Isolate special deletion that fuses rIIA and rIIB
is functional!
Determine if A-B fusion still functional
Mutagenize with proflavin or base-changing
chemicals
a-B (by base-change)
a-b- (by proflavin)
And they found that mutations in rIIB did not
effect rIIA