Chapter 12. Protein biosynthesis - PowerPoint PPT Presentation
1 / 51
Title:
Chapter 12. Protein biosynthesis
Description:
Title: Chapter 12. Protein biosynthesis (P215, sP875) Last modified by: Created Date: 5/27/2004 12:36:25 PM Document presentation format – PowerPoint PPT presentation
Number of Views:244
Avg rating:3.0/5.0
Title: Chapter 12. Protein biosynthesis
1
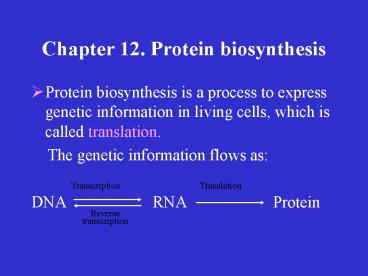
Chapter 12. Protein biosynthesis
- Protein biosynthesis is a process to express
genetic information in living cells, which is
called translation. - The genetic information flows as
- DNA RNA
Protein
Transcription
Translation
Reverse transcription
2
1. Components of Protein Biosynthesis
- Protein biosynthesis requires amino acids, mRNA,
tRNA, ribosomes, protein factors, and synthetic
enzymes. - 1) Messenger RNA a template for protein
biosynthesis, which is read in a 5?3 direction.
Each three nucleotides form a codon representing
for a specific amino acid. Thus, the base
sequence of an mRNA molecule determines the amino
acid sequence of the protein.
3
- Codons in mRNA
4
- mRNA in eukaryotes is usually monocistronic one
mRNA encodes only a single polypeptide chain. - mRNA in prokaryotes usually encodes more than one
polypeptide chain. This is called polycistronic.
5
- Degeneracy of codons refers to the fact that an
amino acid has more than one codon. - one of the consequences of degeneracy is that a
mutation which produces a base change in DNA may
not result in an amino acid change in the encoded
protein. - Synonyms refers to the codons for the same amino
acid. e.g. GUU, GUC, GUA, GUG represent for Val.
6
- B) Universility of codons this genetic code
system is used by all living organisms except in
some cases - in cytosol in
mitochondria - AUA Ile Met
- UGA Stop Trp
- AGA Arg Stop (animal)
- CGG Arg Trp (plant)
- CUN Leu Thr (yeast)
7
- C) Reading frames refer to the different
combinations for each three nucleotides that are
read as a codon each mRNA sequence can be read
in three possible reading frames. - Reading frame 1 UUA UGA GCG CUA AAU
- Leu Stop Ala Leu Asn
- Reading frame 2 U UAU GAG CGC UAA AU
- Tyr Glu Arg Stop
- Reading frame 3 UU AUG AGC GCU AAA U
- Met Ser Ala Lys
8
- D) Open reading frames refer to the runs of
codons that start with ATG and end with TGA, TAA,
or TAG. The open reading frames can be used to
predict the protein sequence encoded.
9
- 2) Transfer RNAs the fidelity of protein
biosynthesis requires tRNAs to serve as adapters
that can recognize the correspondent codons and
carry amino acids to the right positions in
translation. - Each tRNA only brings with it an amino acid, and
recognizes and binds to a specific codon.
10
Secondary structure of tRNA
T?C loop
extra arm
DHU loop
Anticodon loop
11
Tertiary structure of tRNA
12
Codon-anticodon interaction by base pairing
tRNA
13
- Wobble base pairing base pairing between the 3
position of the codon and 5 position of the
anticodon may occur by a non-standard way. This
allows one tRNA to recognize more than one codon.
Examples of wobble base pairing Anticodon wobble
position base C A G U I Codon wobble
position base G U C A C
U G U
A
14
Wobble base pairing of inosine with three
nucleosides
15
- 3) rRNAs and ribosomes As the site of protein
biosynthesis, ribosome is made up of two
subunits, one is large and another is small.
16
Composition of ribosomes in eukaryotes and
prokaryotes
Eukaryotic ribosome (80S)
Prokaryotic ribosome (70S)
large subunit small subunit
large subunit small subunit
Subunit size 60S
40S 50S 30S
rRNAs 5S, 5.8S, 28S 18S
5S, 23S 16S
Proteins 49
33 35
21
17
- A) Polysomes several ribosomes bind to and
translate a single mRNA molecule simultaneously - B) Free ribosomes ribosomes occur free in the
cytosol, usually synthesizing proteins of
cytosol, nucleus, mitochondria or other
organelles - C) Membrane bound ribosomes ribosomes bind to
the membrane of rough endoplasmic reticulum,
usually synthesizing secretory proteins or
membrane proteins.
18
- Polysomes
19
- 4) Aminoacyl-tRNA synthetase is also called
amino acid activating enzyme, which catalyzes the
following reactions.
20
2. Steps of Protein Biosynthesis
- The steps of protein biosynthesis include
initiation, elongation, and termination or
release. - 1) Initiation Translation begins with the
assembly of an initiation complex consisting of
an mRNA, a ribosome, and the initiator tRNA
(fMet-tRNAiMet or Met-tRNAiMet ) . The process
requires a number of protein factors, known as
initiation factors.
21
Formation of the initiation complex in
eukaryotic translation
22
- In prokaryotes, initiation factors IF1 and IF3
bind to the 30S subunit while IF2 binds to
GTPfMet-tRNAiMet. The two complexes and mRNA
combine to form a pre-initiation complex,
releasing IF3. The 50S subunit binds with this
complex, with hydrolyzation of the bound GTP to
GDP and Pi, and release of IF1 and IF2, to form a
completed initiation complex.
23
Formation of the initiation complex in
prokaryotic translation
24
Prokaryotic and eukaryotic initiation factors
Prokaryotic eukaryotic
Function
IF
eIF
IF
binds to small subunit before mRNA bi
nding. eIF
assists
1
1
1
1
mRNA binding.
IF
eIF
eIF
eIF
Bind initiator tRNA, stabilize ternary
complex, cause GTP/GDP
2
2a
2b
2c
exchange.
IF
eIF3 Bind to the
small subunit, assist mRNA binding, cause
3
dissociation of subunits after tr
anslation.
eIF
eIF
eIF
Recognize and bind the mRNA cap, assist mRNA
binding,
4a
4b
4c
eIF
eIF
eIF
hydrolyze ATP to drive scanning for the
initiator codon.
4d
4e
4f
eIF
Promotes GTP hydrolysis and
release of other initiator factors.
5
eIF
Assists subunit dissociation.
6
25
- 2) Elongation Elongation of polypeptide chain
consists of a series of cycles, called ribosomal
cycles, each of which forms a new peptide bond. - Three steps entry, peptide bond formation, and
translocation.
26
A) Entry of aminoacyl-tRNA to the A site of
ribosome (A. in prokaryotes, B. in eukaryotes. AA
aminoacyl)
27
- B) Peptide bond formation
dipeptidyl-tRNA
Peptidyl- transferase
28
- C) Translocation Translocation is a process
involves the shift of the newly formed
peptidyl(n1)-tRNA from the A site to the P site,
with release of the deacylated tRNA from the
ribosome. This process is mediated by another
elongation factor, EF-G in prokaryotes, or eEF2
in eukaryotes.
29
The translocation step in protein biosynthesis
30
- D) Termination when a ribosome moves onto the
stop codon of mRNA, the stop codon in the A site
cannot be recognized by any aminoacyl-tRNA
molecules. Instead, release factors interact with
the mRNA-ribosome complex, leading to discharge
of the newly synthesized polypeptide from the
complex.
31
Termination of protein biosynthesis
32
Elongation and termination factors in prokaryotes
and eukaryotes
33
(Eukaryotic)
34
3. Posttranslational Processing
- Newly synthesized polypeptides usually undergo
structural changes called posttranslational
processing. - The most important posttranslational processing
modification and folding.
35
- Posttranslational modification
- A) Modification of protein primary structures
- Removal of the N-terminal Met residue
36
- Posttranslational processing of human
preproinsulin
37
- B) Glycosylation occurs in most membrane and
secretary proteins, such as glycoproteins. - Two types of glycoproteins in humans O-linked
and N-linked. Formed in endoplasmic reticulum and
Golgi apparatus. - N-linked
O-linked
38
- C) Modification of protein on higher-level
structures - Acetylation of the amino terminus
- Acetyl-SCoA H2N-protein
Acetyl-NH-protein HSCoA - Phosphorylation
- ATP
ADP - Protein
kinase - Protein
Phosphoprotein -
Phosphatase - Pi
H2O
39
- 2) Folding of newly synthesized polypeptides
- Newly synthesized polypeptide chains usually
undergo folding, a process that requires protein
factors called molecular chaperones. Two types of
molecular chaperones chaperones and chaperonins.
- The major function of molecular chaperones is to
assist the correct folding of nascent polypeptide
chains by blocking their hopeless entangling or
insignificant intermolecular interactions.
40
- Molecular chaperones belong to the heat-shock
protein (HSP) family. - Chaperone proteins include HSP70, HSP40, and
GrpE. - The binding-release cycle of chaperone proteins
with a nascent polypeptide earns time for the
proper folding of the unfolded polypeptide chain.
The cycle continues until the polypeptide chain
is folded to a native conformation.
41
The binding-release cycle of a chaperon-polypeptid
e complex
42
- B) Chaperonins are also heat-shock proteins.
They participate in the folding of a variety of
proteins by forming a cylindrical structure (a
ring) enclosing a central cavity. - The target polypeptide chain enters the central
cavity of the folding machine, where it is
properly folded and is then released. The
entering-folding process repeats until a native
3D structure of the protein is formed.
43
A folding cycle of a polypeptide by GroEL-GroES
chaperonins in E. coli cell
44
4. Protein targeting
- Protein targeting is a process in which a newly
synthesized protein is delivered to a specific
extracellular or intracellular location. - Secretory proteins are first synthesized by
ribosomes bound to the rough ER (RER), with a
signal sequence (or called signal peptide) at the
N-terminal end, which directs the protein to be
delivered to its functioning place.
45
Signal peptidase cleavage site
A.
Hydrophobic area
N-
Secretory protein
Signal peptide
Internal signal peptide
B.
N-
Type III integral membrane protein
Signal peptide
Stop-transfer sequence
- Signal peptides of secretory proteins. (B) Type
III integral - membrane proteins with signal-peptide, internal
signal-peptide, - and stop-transfer sequences.
46
- The signal peptide directs a newly synthesized
secretary protein to enter into the RER lumen
SRP signal recognition particle
47
5. Clinical correlation of protein biosynthesis
- Protein biosynthesis is the means to express the
genes that control metabolisms in cells. Any
mistake occurs in protein biosynthesis may result
in severe consequences in metabolism.
48
- Molecular Diseases refer to those resulting from
abnormal protein structures due to mutation of
genes. - Sickle-cell anemia a result from the replacement
of an amino acid residue at position 6 of the
b-chain, glutamate , by another one, valine. - Position of b-chain 1 2 3 4
5 6 7 8 - Hemoglobin A Val-His-Leu-Thr-Pro-Glu-Gl
u-Lys- - Hemoglobin S Val-His-Leu-Thr-Pro-Val-G
lu-Lys-
49
- 2) Action of some antibiotics they carry out
antimicrobial activities via inhibition of
protein synthesis in the microorganism, such as
tetracyclines, streptomycin, chloramphenicol, and
so on.
50
Some antibiotic inhibitors of protein
biosynthesis
51
- 3) Effect of some biological molecules
Interferons (IFNs) are cytokines produced during
immune response to antigens, especially to viral
infections. - Two functions of IFNs cause viral RNA
degradation and inhibit protein biosynthesis in
cells. - IFN protein kinase
phosphorylation of eIF-2a inhibition
of protein biosynthesis in cells
inhibition of the viral replication.