Protein structure prediction - PowerPoint PPT Presentation
1 / 61
Title:
Protein structure prediction
Description:
PSI-PRED secondary structure prediction based on PSIBLAST ... Strand(E), Helix(H) and Coil(C) AA: RLMPHIKRSAIPVNHGQCRWEDNVDERTNCMIQYVLIMRD ... – PowerPoint PPT presentation
Number of Views:110
Avg rating:3.0/5.0
Title: Protein structure prediction
1
Protein structure prediction
Einat Granot Liron Atedgi
2
Protein folding
- Protein folding determined by AA sequence
- Why knowing the folding is importance ?
- Determine its functionality
- Find distant evolutionary relationship
- Design drugs
3
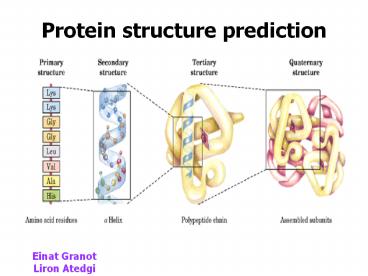
Protein structures
- Primary structure
- Secondary structure
- Tertiary structure
4
Two prediction methods
- PSI-PRED secondary structure prediction based
on PSIBLAST - GenTHREADER tertiary structure prediction
Were developed by the group of David
T.Jones,University of Warwick
5
Methods general format
- Sequence
- Alignment
- Additional
- data
Neuron networks
Structure prediction
6
Neuron networks
7
Neuron networks
Output
Numerical inputs
Units
Why do we call it neuron network ?
Every unit performs weighted calculation
8
Neuron network hidden layer
with the increasing number of added layers the
mean square error is lower
Hidden layer
9
Neuron networks training
- Network connections and weights determined by
training process - Training performs by samples of input and
expected output. - The learning algorithm is called back propagation
10
Network training testing
- After training we perform testing
- Training and testing groups must be chosen very
carefully - What problems can arise ?
- Insufficient training or testing
- Testing group may be biased
11
Neuron networks is a black-box
- The specific algorithm ofa working neuron
networkis not known - Its hard to deduce new biological principles
about the solved problem
12
PSI-PRED Secondary structure prediction
13
Secondary structure prediction
- In DSSP 8 secondary structures categories
- In PSI-PRED were joined into 3Strand(E),
Helix(H) and Coil(C)
AA RLMPHIKRSAIPVNHGQCRWEDNVDERTNCMIQYVLIMRD Pre
d CCCCCHHHCCCCCCEEEEEECCCCCCHHHHEEEEEECCCC
14
PSI-PRED
- sequence alignment (Find homologous)
- Create protein profile
- Insert to first neuron network
- Insert to second neuron network
- Final prediction
15
sequence alignment
- Finding homologous for target protein using
PSI-BLAST - Reminder ? What is PSI-BLAST?
- Position Specific Iterated Blast,giving
output to PSSM.
16
PSI-BLAST Pros Cons
- Pros
- Sensitive to distant homologous
- Reliable
- Accessible from every workstation
- Cons
- Sensitive to distant homologous - Result might be
biased - Sensitive to repetitive sequences
17
Solving PSI-BLAST problems
- A special DB of 340,000 sequences was constructed
for PSI-PRED - This DB contains only unique and unrepetitive
sequences
18
PSI-PRED
- sequence alignment (Find homologous)
- Create protein profile
- Insert to first neuron network
- Insert to second neuron network
- Final prediction
19
Create protein profile
- PSI-PRED uses the PSSM from PSI-BLAST produced
after 3 iteration - This matrix is processed by transformation
- f(x) , so the final values are
- between 0 to 1
20
PSSM Output of PSI-BLAST
Transformation
21
Create protein profile
- The matrix size is M x 20, when M is the sequence
length - Addition column is added which defined the N/C
terminus -gt M x 21 matrix
22
PSI-PRED
- sequence alignment (Find homologous)
- Create protein profile
- Insert to first neuron network
- Insert to second neuron network
- Final prediction
23
Networks training testing
- 187 proteins were selected according to CATH and
PSI-BLAST - CATH filters proteins according to their folding
domains configuration (T-level) - This considered to be a strict selection
24
First neuron network
Every time, a sequence of 15 AA long is inserted
into the first network
E H C
0.2 0.5 0.8
0.1 0.2 0.9
0.8 0.4 0.9
0.2 0.5 0.3
0.7 0.8 0.3
0.2 0.1 0.5
0.3 0.8 0.4
0.9 0.8 0.1
0.2 0.2 0.6
0.7 0.3 0.6
0.3 0.6 0.8
0.3 0.1 0.8
0.7 0.4 0.4
0.6 0.6 0.1
0.4 0.3 0.6
The output is a matrix 15 x 3
25
PSI-PRED
- sequence alignment (Find homologous)
- Create protein profile
- Insert to first neuron network
- Insert to second neuron network
- Final prediction
26
Second neuron network
The input for the 2nd network is the output from
the 1st one
N/C E H C
0 0.2 0.5 0.8
0 0.1 0.2 0.9
0 0.8 0.4 0.9
0 0.2 0.5 0.3
0 0.7 0.8 0.3
0 0.2 0.1 0.5
0 0.3 0.8 0.4
0 0.9 0.8 0.1
0 0.2 0.2 0.6
0 0.7 0.3 0.6
0 0.3 0.6 0.8
0 0.3 0.1 0.8
0 0.7 0.4 0.4
0 0.6 0.6 0.1
1 0.4 0.3 0.6
E H C
0.1 0.5 0.9
0.2 0.1 0.9
0.3 0.4 0.8
0.2 0.7 0.1
0.5 0.8 0.2
0.2 0.7 0.3
0.2 0.9 0.3
0.6 0.8 0.3
0.1 0.3 0.7
0.4 0.2 0.8
0.3 0.5 0.9
0.3 0.2 0.8
0.9 0.2 0.1
0.8 0.3 0.1
0.8 0.3 0.4
Again, another column is added, indicates the N/C
terminus
27
Why do we need a second network?
- Lets examine a possible prediction from
- the 1st network
- What is the problem with this prediction ?
Seq VLFLNDNLDDVVIGRPKRTYTAITL Pred
EEEECCCCHHHCCCHCCCEEEECC
A single AA helix does not exist
The 2nd network maintains the coherency between
adjacent AA and improves the accuracy
28
PSI-PRED
- sequence alignment (Find homologous)
- Create protein profile
- Insert to first neuron network
- Insert to second neuron network
- Final prediction
29
Final prediction
Image of prediction
Degree Of confidence
Target sequence
Secondary structure
30
PSI-PRED evaluation
- CASP Critical Assessment of technique for
protein Structure Prediction experiments - At CASP3 PSI-PRED achieved the best results from
all other methods participated
31
PSI-PRED evaluation
Q3 average PSI-PRED - 76.3 JPRED
72.4 DSC - 67.3
Q3 score percentage of AA predicted correctly
32
Reasons for success
- The use of PSI-BLAST
- More sensitive (iterative algorithm)
- More accurate (pairwise local alignments)
- Usage of neuron networks
- Strict selection for training testing
33
Possible improvements
- Larger data bases (training alignment)
- Combinations with other methods (JPRED)
- Predict more than 3 secondary structure
34
Bring out the food
35
GenTHREADER Tertiary structure Prediction
36
Threading methods
- Trying to thread a target AA sequence on a
template 3D structure
N
S
Q
M
V
D
L
I
R
E
R
A
Q
T
V
L
C
N
K
37
Templates collection
- Target sequence is compared against a collection
of sequences with known folding - The collection was taken from Brookhaven Protein
Data Bank and includes unique sequences
38
GenTHREADER
- Sequence alignment
- Calculate threading potential
- Insert to neuron network
- Final prediction
39
Sequence alignment
- The target sequence is aligned against each of
the templates twice - Target profile against template sequence
- Target sequence against template profile
- The best result is taken
40
Creating a profile
- Steps for creating a profile
- Alignment against OWL DB(A DB for coding
sequences) - Selection of sequences with E-Value lower than
0.01 - Constructing a profile using BLOSUM50
41
Creating a profile
A L M P H I K R S A I P V N H G Y V I M Q C R W E
D N S T K V
42
GenTHREADER
- Sequence alignment
- Calculate threading potential
- Insert to neuron network
- Final prediction
43
Calculate threading potential
- Threading potential includes
- pairwise potential
- solvation potential
44
Pairwise potential
- Potential for interaction between two AA
- Considerate analysis of known structure and
favorable energy configuration - Lower pairwise potential indicates a favorable
state
45
Solvation potential
- Calculated per AA and proportional to its degree
of burial - Degree of burial (DOB) The num of other AA
located in a radius of 10Å - Hydrophobic acids - a high DOB is preferred
- Hydrophilic acids - a low DOB is preferred
46
GenTHREADER
- Sequence alignment
- Calculate threading potential
- Insert to neuron network
- Final prediction
47
Insert to neuron network
- Prediction is very complex therefore a neuron
network is used
48
Neuron network
- Again, the 6 input parameters were converted to
values between 0 1 using the function f(x) - The output is a value between 0 -1 showing the
confidence of the match
49
Network training testing
- The network was trained using pairs of proteins
with known folding patterns - Again the training and testing sets were
separated to avoid bias
50
GenTHREADER
- Sequence alignment
- Calculate threading potential
- Insert to neuron network
- Final prediction
51
Final prediction
- Example for GenTHREADER results
52
GenTHREADER evaluation
- Evaluated using Fischer benchmarking, 68 hard for
predictions proteins pairs - 73.5 were properly detectedBest method - 76.5,
Other methods 50-60 - For the unrecognized proteins the scores were
less than 0.5
53
Genome analysis example
- As an example for GenTHREADER efficiency
Mycoplasma Genitalium genome was analyzed - M.Genitalium is the smallest known bacterial
genome contains 468 ORFs
54
Genome analyzing example
In one day the whole genome was analyzed
Confidence categories
Distribution of protein domain architecture
55
Genome analyzing example
GenTHREADER succeed to predict 46 from the
ORFs. For comparison, in other methods Fischer
Eisenberg 22 Using PSI-BLAST OWL 30
Huynen 38
56
Genome analyzing example
- While analyzing M.Genitalium genome ORF MG353 was
assigned to 1HUE - (Histone like protein)
- ORF MG353 function wasnot known
57
Genome analyzing example
- The results show high similarity in both
secondary structure and functional AA
58
GenTHREADER advantages
- Fast
- No need for human intervention
- Can distinguish false positive in high accuracy
59
Possible improvements
- Improvement of sequence alignment (PSI-BLAST)
- Additional input of sequence features (for
example, secondary structure) - Larger DB of known folding proteins
- Combination with other methods
Already exists (mGenTHREADER)
60
Dont try this at home
- www.psipred.net
- The web is for both GenTHREADER PSI-PRED
61
References
- Jones DT. Protein secondary structure prediction
based on position-specific matrices. J Mol Biol.
1999 292195-202 - Jones DT. GenTHREADER an efficient and reliable
protein fold recognition method for genomic
sequences. J Mol Biol. 1999 287797-815