Coordination Complexes and Transition Metals in Action - PowerPoint PPT Presentation
1 / 60
Title:
Coordination Complexes and Transition Metals in Action
Description:
Normally ligands are anions or polar molecules. Anion (CN-) Polar molecule (NH3) ... Anion = nitrate. Nomenclature ... anion = trioxalatevanadate. Nomenclature ... – PowerPoint PPT presentation
Number of Views:4190
Avg rating:5.0/5.0
Title: Coordination Complexes and Transition Metals in Action
1
Coordination Complexes and Transition Metals in
Action
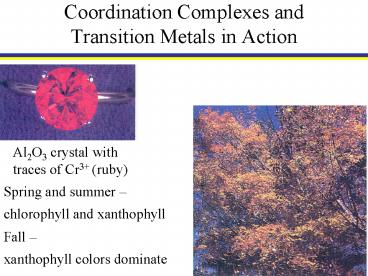
Al2O3 crystal with traces of Cr3 (ruby)
Spring and summer chlorophyll and xanthophyll
Fall xanthophyll colors dominate
2
Plants and Animals
chlorophyll
heme
3
Colors of Chromium
Cr(NO3)3
CrCl3
K2CrO4
K2Cr2O7
Cr3
Cr6
4
Coordination Compound and Complex
Coordination Compound is Co(NH3)6Cl3
- Co(NH3)6Cl3 ? Co(NH3)63 3 Cl-
Coordination Complex is Co(NH3)63
5
Components of Complex (Coordination Sphere)
Co(NH3)63
- Metal ion usually transition metals with empty
valence orbitals - Specifically empty d orbitals
- Act as Lewis acid (electron pair acceptor)
- Ligand complexing agent bound to (surrounding)
the metal ion (Lewis base) - Normally ligands are anions or polar molecules
- Anion (CN-)
- Polar molecule (NH3)
- Donor atom
6
Characteristics of Complex (Coordination Sphere)
- Metal ion
- Oxidation number
- Co ?
- Ligand
- Charge on ligand
- NH3 ?
Charge of Complex sum of charges on the central
metal ion and the surrounding ligands
What is the charge of the complex?
Co(NH3)6Cl3
Coordination number The number of donor atoms
attached to the metal.
7
Example Problem
- Indicate the coordination number of the metal and
the oxidation number of the metal in each of the
following complexes - Na2CdCl4 Co(NH3)4Cl2Cl
- K2MoOCl4 Zn(en)2Br2
8
Types of Ligands
- Monodentate ligand NH3, H2O, Cl-
- Bidentate ligand ethylenediamine (en)
- Polydentate ligand ethylenediaminetetraacetate
ion (EDTA)4-
9
Chelating Agent
- Polydentate ligands (including bidentate) are
called chelating agents because they appear to
grasp the metal between donor atoms
10
11
(No Transcript)
12
Example Ligands
What kind of ligands are these examples?
13
Chelate Effect
- Chelating agents form more stable complexes with
metal ions than monodentate ligands - Ni2 (aq) 6NH3 (aq) ? Ni(NH3)62 (aq) Kf 4
x 108 - Ni2 (aq) 3 en (aq) ? Ni(en)32 (aq) Kf
2 x 1018 - Sequestering agents because the chelating
agents can be used to remove or separate ions - removal of ions from hard water
- removal of trace metals from food
- removal of heavy metal ions from blood
14
Geometry of Complex
- Common coordination of 4
square planar Transition metal ions with 8 d
electrons
Tetrahedral Most common
15
Geometry of Complex
- Coordination 6
Octahedral
16
Effect of Ligand on Coordination Number
- The larger the size of the ligand, the fewer
ligands that can get close enough to bind to the
central metal. - FeF63- FeCl4-
- Ligands which provide negative charge to the
complex reduce the coordination number. - Ni(NH3)62 Ni(CN)42-
17
Metal Complexes
- Distinct chemical properties different from the
metals and ligands from which they were formed. - Different colors
- Different electrochemical properties
- Different solubility properties
18
Nomenclature
- In naming salts, the name of the cation is given
before the name of the anion. - Mo(NH3)3Br3NO3
Cation
Anion nitrate
19
Nomenclature
- Within a complex ion or molecule the ligands are
named before the metal. Ligands are listed in
alphabetical order, regardless of charge on the
ligand. Prefixes that give the number of ligands
are not considered part of the ligand name in
determining alphabetical order. - Mo(NH3)3Br3NO3
Ammonia, bromide, molybdenum
20
Nomenclature
- The names of the anionic ligands end in the
letter o, whereas neutral ones ordinarily bear
the name of the molecules. - Mo(NH3)3Br3NO3
Example ligand names NH3 ammine CO carbonyl
NO - nitrosyl H2O aqua CN- cyano en -
ethylenediammine
Ammine, bromo, molybdenum
21
Nomenclature
- Greek prefixes (di, tri, tetra, penta, hexa) are
used to indicate the number of each kind of
ligand when more than one is present. - Mo(NH3)3Br3NO3
- If the ligand itself contains a prefix, then
these prefixes are used for the ligand name
(bis-, tris-, tetrakis-, pentakis-, etc.). - Ru(bipy)3Cl3
triamminetribromomolybdenum
tris-bipyridineruthenium
22
Nomenclature
- If the complex is an anion, its name ends in
ate. - K3V(C2O4)3
cation potassium
anion trioxalatevanadate
23
Nomenclature
- The oxidation number of the metal is given in
parentheses in Roman numerals following the name
of the metal. - Mo(NH3)3Br3NO3
- Ru(bipy)3Cl3
- K3V(C2O4)3
triamminetribromomolybdenum(IV) nitrate
tris-bipyridineruthenium(III) chloride
Potassium trioxalatevanadate(III)
24
Isomers
Hydrate isomer
Ionization isomer
25
Structural Isomerism have the same numbers and
kinds of atoms, but differ in the bonds that are
present.
- Ionization isomer exchange of ligand with an
anion or neutral molecule
CoBr(NH3)5SO4 and CoSO4(NH3)5 Br - Hydrate isomer the exchange of H2O molecule
with another ligand CrBr(H2O)6Cl3 and
CrClBr(H2O)5Cl2H2O - Coordination isomers- differ due to the exchange
of one or more ligands between a cationic complex
and an anionic complex
Cr(NH3)6Fe(CN)6 - Fe(NH3)6Cr(CN)6
- Linkage isomers- contain the same ligand
coordinated to the metal through different donor
atomsPd(NH3)3SCN and Pd(NH3)3NCS
26
Linkage Isomerism
Nitro
Nitrito
27
Stereoisomerism
- Geometric isomers have the same number and kinds
of bonds, but differ in the relative positions of
the ligands. - Optical isomers rotate the plane of polarized
light in opposite directions.
28
Geometric Isomers
29
Geometric Isomers
30
Optical Isomers
- Rotate the plane of polarized light in opposite
directions.
Levrorotatory rotate left
Dextrorotatory rotate right
31
Optical Isomers
- Chiral molecules have mirror-image structures
that cannot be superimposed. - Only chiral molecules are optically active.
- Enantiomers are chiral molecules of each other.
Racemic Mixture occurs when equal amounts of each
enantiomer are mixed. When this happens, the
optical activity of each is canceled by the other.
32
Enantiomers
33
Properties of Coordination Complexes
- Color Many coordination complexes exhibit a
wide variety of colors, that depend on the metal,
its oxidation state, and the ligands present. - The observed colors result from the absorption of
light in the visible region by the complexes.
Color Exhibited
Colorless
Partially filled d orbital
Totally filled or empty d orbital (d0 and d10)
34
Color Exhibited or Colorless
- Color Exhibited
- Cr(NH3)63
- Fe(SO4)(H2O)4
- Colorless
- Cd(NH3)4(NO3)2
- NaAlCl4
35
Color of Complexes
36
Crystal Field Theory
- Crystal field theory assumes electrostatic
interactions between the negative or neutral
ligands and the positive metal ion lower the
energy of the system. - Anionic ligands electrostatic attraction
- Neutral ligands ion dipole
37
Lowered Energy of Metal/Ligand Complex
Crystal Field repulsive interaction between
electrons in the ligands and the d orbital
electrons in the metal
38
Consequenced electron repulsion
- Crystal Field
- The negative ligands repel the electrons in the
metal ion d orbitals. - The repulsion energy of d electrons depends on
the orientation of the orbital, relative to the
location of the negative ligands.
39
d Orbitals
40
d Orbitals
41
Octahedral complex
42
d orbital splitting
D Energy gap the energy necessary for an
electron to move across the gap is similar to
energy of a visible light photon
Explains why d0 or d10 transition elements do not
show color
43
Spectrochemical Series
44
Electron Configurations
Co3 a d6 ion
45
Electron Configurations
- High spin
- Low spin
Weak Field ligand
Spin pairing energy is the energy required to
pair 2 electrons in orbital
Strong Field ligand
46
Properties of Coordination Complexes
- Paramagnetism - a property due to unpaired
electrons, is common among transition metal
complexes. - Different complexes of the same metal ion, may
have different numbers of unpaired electrons. - Predict the magnetic properties of
- Fe(H2O)62 Fe(CN)64-
47
Energy Calculation
- The complex Ti(H2O)63 absorbs light of
wavelength 510 nm. What is the crystal field d
orbital splitting energy (D) for the complex?
48
Metal and Oxidation
- Color Many coordination complexes exhibit a
wide variety of colors, that depend on the metal,
its oxidation state, and the ligands present. - Cr(H2O)63 V(H2O)62
The larger the charge on the metal ion involved
in the complex, the more metal-ligand
interaction. Therefore, D will be larger when
the oxidation state of the metal is larger.
49
Increased Interaction Between Metal and Ligand
Increased Crystal Field
Crystal Field repulsive interaction between
electrons in the ligands and the d orbital
electrons in the metal
50
Crystal Field Theory Tetrahedral
Tetrahedral complex always have high spin because
D is small
51
Crystal Field Theory Square Planar
Square planar complexes always have high spin
because D is large
52
Tetrahedral and Square Planar
- Draw the crystal field splitting diagrams for
Ni(CN)42- and NiCl42- and predict the
magnetic properties of each.
53
Electron Configurations
- High spin
- Low spin
Weak Field ligand
Spin pairing energy is the energy required to
pair 2 electrons in orbital
Strong Field ligand
54
Energy Calculation
- The complex Ti(H2O)63 absorbs light of
wavelength 510 nm. What is the crystal field d
orbital splitting energy (D) for the complex? - c ln E hn
- c/l E (6.63 x 10-34 Js) (5.88 x
1014 s-1) - 3.00 x 108 m/s______ E 3.90 x 10-19 J
- (510 nm) ___1 m___ E (3.90 x 10-19
J) _1 kJ_ - 1 x 109 nm 1000 J
- 5.88 x 1014 s-1 E 3.90 x 10-22 kJ/photon
- E (3.90 x 10-22 __kJ__) (6.022 x 1023 photons)
235 _kJ_ - photon mole
mole
55
Spectroscopy
56
(No Transcript)
57
Sources of Electromagnetic Radiation
58
Window Material
59
Wavelength Selector
60
Detectors