Diapositiva 1 - PowerPoint PPT Presentation
1 / 20
Title:
Diapositiva 1
Description:
Mara Ercolano (Univ. of Naples) Silvana Grandillo (IGV-CNR, Portici,Naples) ... Mara Ercolano. Sara Melito. Rosa Paparo. Walter Sanseverino. Sara Torre ... – PowerPoint PPT presentation
Number of Views:75
Avg rating:3.0/5.0
Title: Diapositiva 1
1
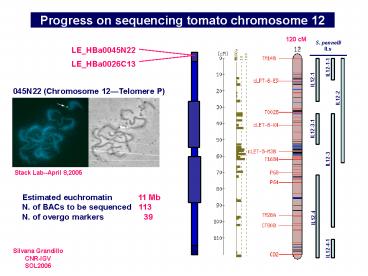
Progress on sequencing tomato chromosome 12
120 cM
S. pennelli ILs
LE_HBa0045N22
LE_HBa0026C13
IL12-1-1
IL12-1
045N22 (Chromosome 12Telomere P)
IL12-2
IL12-3-1
IL12-3
Stack Lab--April 8,2005
Estimated euchromatin 11 Mb N. of BACs
to be sequenced 113 N. of overgo markers 39
IL12-4
IL12-4-1
Silvana Grandillo CNR-IGV SOL2006
2
Tomato chromosome 12
Principal Investigators Luigi Frusciante (Univ.of
Naples) Giovanni Giuliano (ENEA, Rome) Giorgio
Valle (CRIBI, Univ. of Padua) Other
Investigators Amalia Barone (Univ. of
Naples) Maria Luisa Chiusano (Univ. of
Naples) Mara Ercolano (Univ. of Naples) Silvana
Grandillo (IGV-CNR, Portici,Naples) Alessandro
Vezzi (Univ. of Padua)
Funding Agronanotech (Italian Ministry of
Agriculture - started October, 2004) FIRB
(Italian Ministry of Research - started October,
2005) EU-SOL (European Commission - started May
2006)
3
Map position of overgo markers used for Chr. 12
Map Position (cM)
Marker
12,5 C2_At4g03280
Map Position (cM)
Marker
14,0 cLPT-6-E9
19,0 TG263
21,0 TG68
24,0 T1487
51,0 T1045
53,0 T1211
32,0 T1481
53,5 CT99
33,0 T0028
54,0 T1093
Near centromere
36,0 T0989
54,5 C2_At5g42740
39,0 T1667
55,0 T1078
41,0 cLET-8-k4
55,5 TG283
56,0 T1622
57,2 P62
57,5 T1451
65,0 T1947
57,7 cLET-8-E15
66,0 TG111
57,8 T1185
68,0 TG394
58,2 SSR20
68,5 TG367
59,0 CT189
71,0 T1266
60,0 SSR124
60,0 SSR44
60,0 cLET-8-G15
86,0 T1676
96,0 T1784
96,0 TG296
97,0 T0882
101,0 T0770
115,0 T1504
120,0 CD2
4
BAC validation procedure
- Single colony picking
- Medium scale DNA preparative
- BAC-end sequencing
- Fingerprinting
- PCR amplification of genetic marker
- Amplicon sequencing and sequence alignment
- BAC physical location tested by mapping in ILs
lines
5
PCR marker development and IL mapping
PCR
1 BAC C12HBa0161H10 2,3 parental
genotypes 4,5 IL lines 6 negative
control
1 2 3 4 5 6
A
C
B
Multiple alignment of S. pennellii, M82, IL
12-2, BAC P161H10, EST T1045 and IL 8-1
S. pennellii T1045 IL8-1 P161H10
IL12-2 M82
Centromeric region
T1045
E
S. pennellii T1045 IL8-1 P161H10
IL12-2 M82
S. pennellii T1045 IL8-1 P161H10
IL12-2 M82
6
Sequencing and assembly strategy
Sequencing template Moving from PCR product to
plasmid mini-preparation Average plasmid insert
size 2000bp Usually double-barrel strategy
(when the overlap between BACs is very high, the
choice has been to sequence only one end of the
plasmid clones, and then to sequence from the
other end only the informative clones) Assembly
program phred/phrap/consed package Sequencing
standard for each BAC lt 3 single strand
region lt 1 single subclone (possibly none) 8-10X
coverage
7
Sequencing and assembly (CRIBI, Univ of Padua)
8
Summary
To date 18 seed BACs associated to 16 markers (11
mapping on the short arm and 5 on the long arm of
chr. 12) have been selected for validation and
sequencing. The map position of the clones is
being confirmed by means of SNPs identified on
the S. pennellii IL population. A total of 23
BACs are currently at different phases of the
sequencing pipeline. 4 seed BAC sequences (Phase
3) have been submitted to SGN repository. In
order to identify new BACs to move out of the 4
finished seed BACs, as well as of the Phase 1 or
2 BAC clones, a program complementary to the SGN
Online BLAST Interface has been developed at
CRIBI (Univ. of Padua). A bioinformatic
platform has been built at the Univ. of Naples
to provide an Italian resource for supporting the
annotation of the tomato genome.
9
Sequencing status of selected BAC clones
LE_HBa0140M01 (C2_At4g03280)
LE_HBa0061F16
LE_HBa0026C13 (cLPT-6-E9)
LE_HBa0090D09
LE_HBa0073O10
LE_HBa0260C13 LE_HBa0206G16 (T1487)
LE_HBa0075C18 (T1481)
LE_HBa0163O04 (T0028) (Map position ???)
SL_MboI0126D24
LE_HBa0032K07 (T0989)
SL_EcoRI0082A18
LE_HBa0180O10 (cLPT-8-K4)
LE_HBa0244C09 LE_HBa0146I19 (T1667)
LE_HBa0161H10 (T1045)
LE_HBa0149G24
LE_HBa0021L02 (T1211)
SL_EcoRI0004H16
LE_HBa0059A05 (SSR124)
LEGEND
LE_HBa0193C03 (T1266)
of BACs
(2)
Validated
LE_HBa0115G22 (T1676)
(5)
In pipeline
HTGS phase 1
(6)
LE_HBa0093P12 (T0882)
HTGS phase 2
(4)
LE_HBa0183M06 (T0770)
(4)
Finished submitted
Finished but problems
(4)
25
LE_HBa0147G13 (T1504 seq TG350)
Extension in progress
BAC for extension at 5 in sequencing pipeline
BAC for extension at 3 in sequencing pipeline
10
Sequencing status of selected BAC clones
11
Sequencing status of selected BAC clones
12
Problems
1. No marker-specific amplification by PCR of few
selected seed BACs
2. Problems with BAC identity LE_HBa0075C18
BES verified, but no marker in the consensus
sequence LE_HBa0260C13 no BES available at
SGN LE_HBa0244C09 no BES available before
sequencing, then identity not confirmed
3. Wrong map position for LE_HBa0163O04 BES
verified, marker enclosed, FISH map on chr. 7,
working on assembly for hard repeat region
4. BACs with repeat regions LE_HBa0059A05
easily resolved LE_HBa0163O04 hard repeat
region ( gt plasmid insert size) LE_HBa0147G13
still to work
5. BAC extention How to move out of a
sequenced BAC? Should we trust a fully automated
BLASTN? How to choose the right BAC?
13
BAC EXTENSION BacEnds Extension v 0.1
developed at CRIBI (Univ. of Padua)
This tool has been designed to simplify some of
the problems commonly found during the BAC
extension procedure in the Tomato Genome
Project. Repetitive sequences may easily act as a
bridge to many BAC ends belonging to a different
part of the genome. The program is already
available to the Solanaceae community
http//tomato.cribi.unipd.it/ USER tomato PWD
trial
For further informations please contact Dott.
Davide Campagna e-mail davide_at_cribi.unipd.it
CRIBI, University of Padua
14
BAC EXTENSION BacEnds Extension v 0.1
developed at CRIBI (Univ. of Padua)
- This tool displays the alignments of a BAC
against the available tomato BAC ends - The
BAC ends are displayed showing their orientation,
thus indicating the direction where the
corresponding BAC is extending - The
electrophoretic profiles can be opened allowing
an immediate control of any discrepancy - The
RAP (Repeat Analysis Program) and Low Complexity
indexes are shown indicating the
repetitiousness of each part of the BAC -
Any sequence found in correspondence to a repeat
should not be considered as reliable for BAC
extension Campagna D. et al. 2005, RAPa new
computer program for de novo identification of
the repeated sequences in whole genomes
Bioinfomatics 21 (5) 582-588
15
BacEnds Extention v 0.1
Sequenced BAC coordinates
RAP index
Low complexity index
BAC end sequences
Developed at CRIBI, University of Padua, by Dott.
Davide Campagna (e-mail davide_at_cribi.unipd.it)
Available at http//tomato.cribi.unipd.it/ USER
tomato PWD trial
16
BacEnds Extention v 0.1
Blast against BACENDS database
Blast results parsering
BAC sequence
Analyse the sequence and Builds RAP index
RAP database
Data Structure
Partial image of BACENDS alignments, RAP and
Linguistic Complexity index
Images of profiles aligned with blast results
17
Bioinformatics at Univ. of Naples
- A bioinformatic platform has been built to
provide an Italian resource to support the
experimental annotation of the Solanum
lycopersicum genome - Annotated EST database from dbEST (NCBI) for
Tomato and Potato species (updated May 2006) - Gbrowse interface for BAC annotation which has
been released to the SOL community - http//biosrv.cab.unina.it
- -The BACs available at the SGN site were
experimentally annotated and are available
through the Generic Genome Browser web interface
allowing selection of reference Gene Models to
test predictive approaches. - The two EST databases and the Gbrowse are
cross-referenced and linked to the Solanaceae
Genome Network resources to provide useful data
integration.
DAgostino, Aversano and Chiusano 2005,
ParPEST A pipeline for EST data analysis based
on parallel computing BMC Bioinformatics, 6
(Suppl. 4)S9
18
(No Transcript)
19
(No Transcript)
20
Acknowledgments
University of Naples Federico II (Dept.
DISSPA) Luigi Frusciante Amalia Barone Mara
Ercolano Sara Melito Rosa Paparo Walter
Sanseverino Sara Torre
ENEA, Rome Giovanni Giuliano Elio Fantini Alessia
Fiore Giuseppe Puglia
CRIBI and Univ. of Padua Giorgio Valle Alessandro
Vezzi Davide Campagna Laura Colluto Michela
D'Angelo Fabrizio Levorin Giorgio Mitch
Malacrida Silvia Pescarolo Riccardo Schiavon Sara
Todesco Alessandro Zambon
University of Naples Federico II (Dept.
DSFB) Maria Luisa Chiusano Mario Aversano Nunzio
D'Agostino Alessandra Traini
CNR-IGV, Portici (Naples) Silvana Grandillo Maria
Cammareri Pasquale Termolino