Course Outline - PowerPoint PPT Presentation
1 / 61
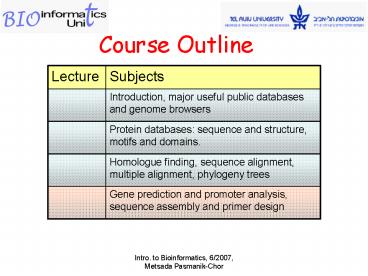
Title: Course Outline
1
Course Outline
2
What Happens when HGP is Completed ?
- Known gene number, location, function and
regulation. - Chromosome structure and organization.
- Non-coding DNA types, amount, distribution and
function. - Gene expression, protein synthesis, and
post-translation events, - protein interactions and networks.
- Evolutionary conservation among organisms,
(structure and function). - Correlation of SNPs with health and disease.
- Genes involved in complex traits and multi-gene
diseases.
3
The Next Step
Locate all the genes and describe their
function. This will probably take another 15-20
years !
4
Reminder Genome Browsers and Gene Prediction
http//genome.ucsc.edu/
5
Eukaryotes vs Prokaryotes
Typical human bacterial cells drawn to
scale.
- Eukaryotic cells are
- characterized by
- membrane-bound
- compartments,
- which are absent
- in prokaryotes.
BIOS Scientific Publishers Ltd, 1999
6
Gene Prediction is Different for Eukaryotes and
Prokaryotes
- Prokaryotic genes
- Eukaryotic genes
http//csbl.bmb.uga.edu/resources/slides/gene-find
ing-2.ppt275,4,Review
7
Eukaryotes Splice Signals
8
The operon model of prokaryotic gene regulation
- Small genomes, high gene density -
- Example Haemophilus influenza genome
- 85 genic.
- Groups of genes coding for related
- proteins are arranged in units known as
- operons.
- No introns - One gene, one protein.
- Open reading frames - One ORF per gene, ORFs
begin with start, end with stop codon.
http//www.emc.maricopa.edu/faculty/farabee/BIOBK/
BioBookGENCTRL.html
9
Gene finding is simple !
http//tardigrada.cap.ed.ac.uk/teaching/genomics/t
ech2/img2.htm
10
Prokaryotic Gene Prediction Tools
- Glimmer
- http//cbcb.umd.edu/software/glimmer/
- GeneMark
- http//exon.gatech.edu/genemark/genemark_prok_gms_
plus.cgi - ORNL Annotation Pipeline
- http//compbio.ornl.gov/GP3/pro.html
- FramePlot - protein-coding region prediction tool
for high GC-content bacteria - http//www.nih.go.jp/jun/cgi-bin/frameplot.pl
11
Gene Prediction in Eukaryotes
- Complex gene structure.
- Large genomes (0.1 to 3 billion bases).
- Exons and introns (interrupted).
- Low coding density (lt30).
- 2-3 in humans, 25 in Fugu, 60 in yeast
- Alternate splicing (40-60 of all genes).
- Considerable number of pseudogenes.
NATURE BIOTECHNOLOGY VOLUME 25 NUMBER 8 AUGUST
2007
12
Gene Prediction in Eukaryotes
13
Non-coding In Genomes
http//fig.cox.miami.edu/cmallery/150/gene/noncod
ing.genes.jpg
14
Non-Protein Coding Gene Tools
- tRNA
- tRNA-ScanSE (http//www.genetics.wustl.edu/eddy/tR
NAscan-SE/) - FAStRNA (http//bioweb.pasteur.fr/seqanal/interfac
es/fastrna.html) - snoRNA
- snoRNA database (http//rna.wustl.edu/snoRNAdb/)
- microRNA
- Sfold (http//www.bioinfo.rpi.edu/applications/sfo
ld/index.pl) - SIRNA (http//bioweb.pasteur.fr/seqanal/interfaces
/sirna.html)
http//harlequin.jax.org/GenomeAnalysis/GeneFindin
g04.ppt257,38,Non-protein Coding Gene Tools and
Information
15
Approaches to Gene Finding
- Direct Look for something that looks like one
gene (homology) - Homology based gene prediction - Exact or near-exact matches (similarity searches)
of - EST, cDNA, or proteins from the same, or closely
related - organisms (comparative genomics).
- Indirect
- Look for something that looks like all genes (ab
initio). - Hybrid
- Combine homology and ab initio.
16
(No Transcript)
17
(No Transcript)
18
General Things to Remember about
(Protein-Coding) Gene Prediction Software
- Work best on genes that are reasonably similar to
something seen previously. - Finds protein coding regions far better than
non-coding regions. First and last exons are
difficult to annotate because they contain UTRs.
Small genes are not statistically significant and
therefore hard to predict. - In the absence of external (direct) information,
alternative forms will not be identified. - It is imperfect ! (Its biology, after all).
19
Finding Eukaryotic Genes Computationally
- Gene finding based on homology evidence BLAST,
FASTA, BLAT etc. - Content-based Methods
- CpG islands, GC content, hexamer repeats,
composition statistics, codon frequencies - Feature-based Methods
- donor sites, acceptor sites, promoter sites,
start/stop codons, polyA signals, feature lengths - Similarity-based Methods
- sequence homology (DNA, protein), EST searches
- Pattern-based
- HMMs, Artificial Neural Networks
- Most effective is a combination of all the above !
20
Content-Based Methods
- CpG islands (in and near approximately 40 of
promoters of mammalian genes (about 70 in human
promoters) - Very abundant near gene start site
- High GC content found in 5 ends of genes
- Codon Bias
- Some codons are strongly preferred in coding
regions, others are not - Positional Bias
- 3rd base tends to be G/C rich in coding regions
21
Feature-Based Methods
- Based on identifying gene signals (promoter
elements, splice sites, start/stop codons, polyA
sites, etc.) - Wide range of methods
- Consensus sequences
- Weight matrices
- Neural networks
- Decision trees
- Hidden Markov Models (HMMs)
22
Eukaryotic Genomic Hints that a Gene is Nearby
- PolII RNA promoter elements
- GC box, TATA box, CCAAT region.
- Kozak consensus sequence (eukaryotic ribosome
binding site-RBS consensus (gcc)gccRccAUGG,
where R is a purine (adenine or guanine) three
bases upstream of the start codon (AUG). - Splicing signals donor, acceptor.
- Termination signal.
- Polyadenylation signal.
- Promoter elements.
23
Pol II Promoter Elements
- 5 cap region/signal
- (a modified guanine nucleotide that has been
added to the "front" or 5' end of a eukaryotic
messenger RNA shortly after the start of
transcription, important for ribosome
recognition). - nCAGTnG
- TATA box (30 bp upstream)
- - TATAAA
- CCAAT box (80 bp upstream)
- - TAGCCAATG
- GC box (200 bp upstream)
- - GGGCGG
- None of these are essential for gene expression
- Each of these may have more than one copy, except
the TATA box, to produce greater effect - There are other promoter elements
24
Control of Gene Expression Promoter Prediction
25
How does it Works Motif Identification
- Exon-Intron Borders Splice Sites
Exon Intron
Exon gaggcatcagGTttgtagactgtgtttcAG
tgcacccact ccgccgctgaGTgagccgtgtc
tattctAGgacgcgcggg tgtgaattagGTaagaggtt
atatctccAGatggagatca ccatgaggagGTgagtg
ccattatttccAGgtatgagacg
Splice site Splice site
Motif Extraction Programs at http//www-btls.jst.g
o.jp/ Tools for genome analysis
http//www-btls.jst.go.jp/cgi-bin/Tools/index.cgi?
langen
http//www.seas.gwu.edu/simhaweb/cs177/spring2004
/zeeberglecture1Part2.ppt317,25,How it works I
Motif identification
26
Prediction of Splice Junction Sites
Splice site prediction tools - 3 splice site
CAG/GT 5 splice
site MAG/GTRAGT
(M is A or C R is A or G). http//l25.itba.mi.cn
r.it/webgene/wwwspliceview.html
http//dot.imgen.bcm.tmc.edu9331/seq-search/gene
-search.html Splice predictor
http//bioinformatics.iastate.edu/cgi-bin/sp.cgi
Splice site prediction by Neural Network
http//www.fruitfly.org/seq_tools/splice.html
27
Prediction of Translation Start Site
- Translation start ATG
- How to predict a translation start
ATG
GCCATGGCGA .. ACGATGCTGT . GACATGGTAC
AGGATGGGCT GCGATGTGGC
AUG codon finder tool http//www.cbs.dtu.dk/
services/NetStart/ http//l25.itba.mi.cnr.it/web
gene/wwwaug.html
28
Polyadenylation Termination Signals
- Polyadenylation signal
- AA/TTAAA
- Located 20 bp upstream of poly-A cleavage site
- Termination Signal
- AGTGTTCA
- Located 30 bp downstream of poly-A cleavage site
29
Why Polyadenylation is Really Useful
Complementary Base Pairing
AAAAAAAAAAA TTTTTTTTTTT
30
Links for Gene Finding Software
POLY-A signal prediction - 3 end of most
eukaryotic mRNAs (60-200 residues),
post-transcription added http//dot.imgen.bcm.tmc
.edu9331/seq-search/gene-search.html
http//l25.itba.mi.cnr.it/genebin/wwwHC_POLYA
http//genomic.sanger.ac.uk/gf/gf.shtml http//ru
lai.cshl.org/tools/polyadq/polyadq_form.html
Repeated elements (Repeat Masker) http//ftp.ge
nome.washington.edu/RM/RepeatMasker.html
http//l25.itba.mi.cnr.it/genebin/wwwrepeat.pl
GC rich areas, http//rulai.cshl.org/tools/CpG_p
romoter/
31
The Annotation Pipeline
- Mask repeats using RepeatMasker
(http//www.repeatmasker.org/). - Run sequence through several gene prediction
programs. - Validation
- Take predicted genes and do similarity search
against ESTs and genes from other organisms. - Do similarity search for non-coding sequences to
find ncRNA.
32
Eukaryotic Gene Prediction Tools
- Genscan (ab initio), GenomeScan (hybrid)
- (http//genes.mit.edu/)
- (http//genes.mit.edu/genomescan.html)
- Twinscan (hybrid)
- (http//genes.cs.wustl.edu/)
- FGENESH (ab initio)
- (http//www.softberry.com/berry.phtml?topicgfind)
- GeneMark.hmm (ab initio)
- (http//opal.biology.gatech.edu/GeneMark/eukhmm.cg
i) - MZEF (ab initio)
- (http//rulai.cshl.org/tools/genefinder/)
- GrailEXP (hybrid)
- (http//grail.lsd.ornl.gov/grailexp/)
- GeneID (hybrid)
- (http//www1.imim.es/software/geneid/geneid.html
) - FirstEF
- - http//rulai.cshl.org/tools/FirstEF/
33
Gene Finding Software Promoter Prediction
Promoter Prediction McPromoter
http//genes.mit.edu/McPromoter.html Human
Promoter Prediction http//www.softberry.com/berr
y.phtml?topicfpromgroupprogramssubgrouppromot
er Consite tool http//asp.ii.uib.no8090/cgi-bi
n/CONSITE/consite?rmt_input_single DNA Motif
search (in TRANSFAC) http//motif.genome.jp/
TFBind http//tfbind.ims.u-tokyo.ac.jp/
TFSearch http//www.cbrc.jp/research/db/TFSEARCH
.html http//molsun1.cbrc.aist.go.jp/research/db/
TFSEARCH.html Promoter prediction - DNA region
that RNA polymerase binds before initiating
transcription (TATA box prediction) http//www.fr
uitfly.org/seq_tools/promoter.html Transcription
factor binding sites prediction http//www.cbil.u
penn.edu/tess/index.html
34
Example - gene finding tool http//genes.mit.e
du/genomescan.html
35
When selecting ORF, you can blast the amino
acid sequence and get a protein.
http//www.ncbi.nlm.nih.gov/gorf/gorf.html
6 frames
36
GENSCAN output for sequence
Optimal exon
Initial exon
Internal exon
Terminal exon
Single Exon gene
Sub-Optimal exon
http//genes.mit.edu/
37
Gene Prediction Pipeline - Example
- Get a genomic sequence from genome browser
- BRCA1 genomic NG_005905 Fasta format.
- Apply to gene prediction tools provided
- GENSCAN http//genes.mit.edu/GENSCAN.html
- ORFinder http//www.ncbi.nlm.nih.gov/gorf/go
rf.html - Also, worth trying
- GeneBuilder http//l25.itba.mi.cnr.it/webgene/g
enebuilder.html - Compare results.
- Try CONSITE example for Transcription factor
recognition of promoter region - (http//asp.ii.uib.no8090/cgi-bin/CONSITE/consite
?rmt_input_single)
Step-by-step practical exercise for gene
annotation http//www1.imim.es/eblanco/seminars/
docs/Reus2005/exercise1/index.html
38
Annotation of Putative Genes
Annotation category
- Matches known protein sequence
- Strong similarity to protein sequence
- Similar to known protein
- Similar to unknown protein
- Similar to EST (i.e., putative protein)
- No EST or protein matches (i.e., hypothetical
protein)
39
Sequencing and Assembling a Genome
- To sequence a genome, the first task is to cut it
into many small, overlapping pieces. - Then clone each piece.
http//media.wiley.com/assets/1312/61/chapter05.pp
t317,15,Sequencing and Assembling a Genome (I)
40
Sequencing and Assembling a Genome
- Each piece must be sequenced.
- Sequencing machines cannot do an entire sequence
at once. - They can only produce short sequences smaller
than 1 Kb. - These pieces are called reads.
- It is necessary to assemble the reads into
contigs.
http//media.wiley.com/assets/1312/61/chapter05.pp
t322,16,Sequencing and Assembling a Genome (II)
41
Cleaning DNA Sequences
- In order to sequence genomes, DNA sequences are
often cloned in a vector (plasmid, YAC, or
cosmide). - Sequences of the vector can be mixed with your
DNA sequence. - Before working with your DNA sequence, you should
always clean it with VecScreen
http//www.ncbi.nlm.nih.gov/VecScreen/VecScreen.ht
ml
42
Gene Assembly
Usually genes are found in fragments, and need to
be assembled Simple example input
ACCGT CGTGC TTAC TACCGT
Output --ACCGT-- ----CGTGC
TTAC----- -TACCGT--
Spaces are ignored. Fragments are
assembled according to overlapping areas.
___________ TTACCGTGC
A multiple alignment of the fragments result in
consensus sequence.
Assembled piece 9 bases
43
Sequence Assembly - Real life is more complicated
Errors in sequencing Example
Output --ACCGT--
----CGTGC TTAC-----
-TGCCGT--
Input ACCFT CGTGC TTAC TGCCGT
TTACCGTGC
The consensus is still correct because of
majority voting.
Insertion error Example
Output
--ACC-GT-- ----CAGTGC TTAC------ -TACC-GT--
Input ACCGT CAGTGC TTAC TACCGT
TTACC?GTGC
44
Gene Assembly - Real life is more complicated
Deletion error Example Input ACCGT Output
--ACCGT-- CGTGC
----CGTGC TTAC TTAC----- TACGT
-TAC_GT--
Repeats
TTACCGTGC
x
x
x
Lack of coverage
Target DNA
Uncovered area
45
Sequence Assembly
CAP3 http//pbil.univ-lyon1.fr/cap3.php http//bi
oweb.pasteur.fr/seqanal/interfaces/cap3.html
http//deepc2.psi.iastate.edu/aat/cap/cap.html
Example sequences (seq) http//genome.cs.mtu.edu/
cap/data/
46
Compute a Restriction Map
NEBcutter V2.0 http//tools.neb.com/NEBcutter2/in
dex.php http//bioinformatics.org/sms2/rest_map.ht
ml WatCut An on-line tool for restriction
analysis, silent mutation scanning, and SNP-RFLP
analysis http//watcut.uwaterloo.ca/watcut/watcut
/template.php Sequence Extractor generates a
clickable restriction map and PCR primer map of a
DNA sequence. Protein translations and
intron/exon boundaries are also shown
http//bioinformatics.org/seqext/
47
Perform PCR Using a Computer
- Polymerase Chain Reaction (PCR) is a method for
amplifying DNA. - PCR is used for many applications, including
- Gene cloning
- Forensic analysis and paternity tests
- PCR amplifies the DNA between two anchors called
PCR primers. - Hydrogen bonding of single-stranded nucleic acids
is referred to as "annealing two complementary
sequences will form hydrogen bonds between their
complementary bases (G to C, and A to T or U) and
form a stable double-stranded molecule.
primers
http//media.wiley.com/assets/1312/61/chapter05.pp
t309,6,Making PCR with a Computer
48
Degenerate Primers
Degenerate Primers are used for amplification of
sequences from different organisms a set of
primers which have a number of options at several
positions in the sequence so as to allow
annealing to and amplification of a variety of
related sequences.
http//www.mcb.uct.ac.za/pcroptim.htm http//www.s
cielo.br/img/revistas/mioc/v100n6/a08fig01.gif
49
PCR Primer Design 10 Rules
1. Primer length should be 18-22 bases in
length This length is long enough for adequate
specificity, and short enough for primers to bind
easily to the template at the annealing
temperature. 2. Base composition determines
DNA stability (40-60 GC content higher GC
makes more stable DNA) 3. GC clamp Primers
should end (3') in a G or C, or CG or GC this
promotes specific binding 4. Runs of three or
more Cs or Gs at the 3'-ends of primers may
promote mis-priming at G or C-rich sequences
(because of stability of annealing), and should
be avoided. http//www.premierbiosoft.com/tech
_notes/PCR_Primer_Design.html
50
PCR Primer Design 10 Rules
5. Primer Tms (melting temp.) between 52-58oC
are preferred definition the temperature at
which one half of the DNA duplex will dissociate
to become single stranded and indicates the
duplex stability. Annealing temp
determines reaction specificity. 6. Secondary
structure 3'-ends of primers should not be
complementary (ie. base pair), as otherwise
primer self dimers will be synthesized
preferentially to any other product 7.
Secondary structure Cross dimers formed
between pairs of primers. 8. Target sequence
Amplicon length (usually 100-500 bps). 9. Check
cross homology with related genes and possible
pseudo-genes. Amplicon location (distance from
3'end). 10. Intron spanning.
http//www.premierbiosoft.com/tech_notes/PCR_Prime
r_Design.html
51
Primer Design Tools
Primer3 http//biotools.umassmed.edu/bioapps/pr
imer3_www.cgi PrimerQuest (also option for
RT-PCR) http//www.idtdna.com/Scitools/Applicatio
ns/PrimerQuest PCR primers based upon
multialignments (PriFi use demo) http//cgi-www.
daimi.au.dk/cgi-chili/PriFi/main?config.x101conf
ig.y30 PCR primers designed from protein
multiple sequence alignments http//bioinformatics
.weizmann.ac.il/blocks/codehop.html GeneFisher
input single or multiple sequence(s), either
nucleotide or amino acid. For multiple sequences
a multiple alignment will be calculated (or
upload already aligned sequences).
http//bibiserv.techfak.uni-bielefeld.de/genefishe
r2/submission.html (example sequences in
web-site).
52
Example
53
Special Features Primer Design
Create overlapping PCR products in large
sequences. http//www2.eur.nl/fgg/kgen/primer/Over
lapping_Primers.html Creating primers around
exons in genomic DNA. http//www2.eur.nl/fgg/kgen/
primer/Genomic_Primers.html Creating primers
around SNPs in genomic DNA. http//www2.eur.nl/fg
g/kgen/primer/SNP_Primers.html Creating primers
around the Open Reading Frame of cDNAs.
http//www2.eur.nl/fgg/kgen/primer/cDNA_Primers.h
tml
54
Examine Primer Properties
NetPrimer http//www.premierbiosoft.com/netprime
r/netprlaunch/netprlaunch.html Blast primer
sequence against NCBI and provide primer
propertieshttp//www.idtdna.com/analyzer/Applica
tions/OligoAnalyzer/ OligoCalculator
Oligonucleotide properties calculator http//www.
basic.northwestern.edu/biotools/oligocalc.html
OligoAnalyzer http//www.idtdna.com/analyzer/Ap
plications/OligoAnalyzer/
55
Other Primer Utilities
UCSC In-Silico PCR UCSC http//genome.ucsc.ed
u/cgi-bin/hgPcr?commandstart Electronic PCR
(e-PCR) is computational procedure that is used
to identify sequence tagged sites(STSs), within
DNA sequences http//www.ncbi.nlm.nih.gov/sutils/
e-pcr/ In silico simulation of molecular
biology experiments http//insilico.ehu.es/
http//insilico.ehu.es/PCR/
56
Primers Design Pipeline
- Use accurate sequence data as much as possible
! - Restrict primer search to regions that best
reflect goals - Locate candidate primers
- Discard candidate primers that show
undesirable self-hybridization - Verify the site-specificity of the primer
57
Primers Design Pipeline (example)
- Question
- Hybrid cells are formed containing human BRCA1
cDNA in mouse cells. - We want to identify cells which had incorporated
the human cDNA, using PCR primers. - Steps
- Look for mouse and human BRCA1 in NCBI/GENE
databse. - Locate BRCA1 cDNA sequence (is there only 1
transcript ?). - If there are more than 1 transcripts, compare
between them (lets take only 3 for this example)
and find the region that would identify all
transcripts (hint use http//www.ebi.ac.uk/Tools
/clustalw/index.html). - Find primer-pairs for PCR (http//frodo.wi.mit.e
du/primer3/input.htm). - Check primer specificity (human only, not
mouse) - UCSC http//genome.ucsc.edu/cgi-bin/hgPcr?comman
dstart and/or - NCBI (BLAST) http//genome.ucsc.edu/cgi-bin/hgPc
r?commandstart - Check primer properties (http//www.basic.north
western.edu/biotools/oligocalc.html)
58
Bioinformatics - Past and Present
ORTHOLOG GENES (Taxonomy)
SEQUENCE ALIGNMENT
CODING REGIONS
CONSERVED DOMAINS
SEQUENCES LITERATURE
3-D STRUCTURE
GENE FAMILIES
SIGNAL PEPTIDE
MUTATIONS POLYMORPHISM
GENOME MAPS
CELLULAR LOCATION
59
Bioinformatics - Present and Future
ORTHOLOG GENES (Taxonomy)
SEQUENCE ALIGNMENT
CODING REGIONS
CONSERVED DOMAINS
GENE EXPRESSION, GENES FUNCTION, DRUG PERSONAL
THERAPY
3-D STRUCTURE
GENE FAMILIES
SIGNAL PEPTIDE
MUTATIONS POLYMORPHISM
GENOME MAPS
CELLULAR LOCATION
60
Are We Done ?
Now this is not the end. It is not even the
beginning of the end. But it is perhaps, the
end of the beginning. Winston Churchill, 1942
(3 years into WW2)
http//www.globecartoon.com/neweconomy/10.html
61
Thank-you !
Dr. Metsada Pasmanik-Chor Bioinformatics Unit,
001 Sherman Bldg. Faculty of Life Science,
TAU Tel x 6992 E-mail metsada_at_bioinfo.tau.ac.il
Bioinfo. Unit webpage http//www.tau.ac.il/lifes
ci/bioinfo/