Mammalian cell genetics - PowerPoint PPT Presentation
1 / 44
Title:
Mammalian cell genetics
Description:
Genetics as a subject (genetic things that go on in somatic cells: ... Recessive mutations (most knock outs) are masked. 10/19/09. 4. Solutions to diploidy problem: ... – PowerPoint PPT presentation
Number of Views:171
Avg rating:3.0/5.0
Title: Mammalian cell genetics
1
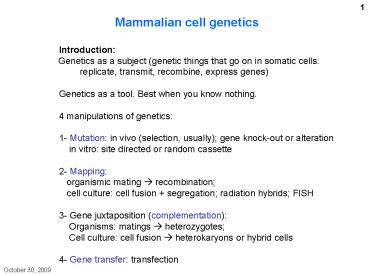
Mammalian cell
genetics Introduction Genetics as a
subject (genetic things that go on in somatic
cells replicate, transmit, recombine, express
genes) Genetics as a tool. Best when you know
nothing. 4 manipulations of genetics 1-
Mutation in vivo (selection, usually) gene
knock-out or alteration in vitro site
directed or random cassette 2- Mapping
organismic mating ? recombination cell
culture cell fusion segregation radiation
hybrids FISH 3- Gene juxtaposition
(complementation) Organisms matings ?
heterozygotes Cell culture cell fusion ?
heterokaryons or hybrid cells 4- Gene transfer
transfection
2
Mammalian cell genetics cont. Advantages of
cultured cells numbers, homogeneity Disadvantag
es of cultured mammalian cells limited
phenotypes limited differentiation in culture
(but some phenotypes available) no sex (cf.
yeast)
Mammalian cell lines Most genetic manipulations
use permanent lines, for the ability to do
multiple clonings Primary, secondary cultures,
passages, senescence. Crisis, established cell
lines, immortality vs. unregulated growth. Most
permanent lines immortalized, plus
"transformed, (plus have abnormal karyotypes)
3
Mutation in cultured mammalian cells Problem of
epigenetic change Variants vs. mutants Stable
heritable alterations in phenotype that are not
due to mutations heritable switches in
gene regulation (?) DNA CpG methylation,
histone acetylation/de-acetylation Diploidy.
Heteroploidy. Haploidy. The problem of diploidy
and heteroploidy (cf. e.g., yeast, or C. elegans,
Dros.) F2 ? homozygotes) Recessive mutations
(most knock outs) are masked.
4
Solutions to diploidy problem Dominant
mutations only (too limited) Haploid cells
hasn't worked just modest chromosome reductions
in CHO Use haploid genes (XY) or functionally
haploid (XX, allelic exclusion)
restricitive Heterozygous loci (rare, despite
CHO reputation) Double mutants (incl. mutation
segregation, or mutation homozygosis(rare but
can be done) Heavy mutagenesis, mutants/survivor
increases but mutants/ml decreases. How hard is
it to get mutants? What are the spontaneous and
induced mutation rates? Measurement of
spontaneous mutation rates. Rate vs. frequency
(freq includes accumulation. Spont
10-7/cell-generation Induced as high
as 10-3 /cell (EMS, UV) Note Same
considerations for creation of double knockout
recessive tumor suppressor genes in cancer.
5
Loss of heterozygosity (LOH)by mitotic
recombination between homologous chromosomes
(rare)
or
-
-
-
-
-
-
Heterozygote
After homologous recombination (not sister
chromatid exchange)
1 homozygote 1 homozygote -
6
Mutagenesis. Chemical and physical agents
MNNG point mutations (single base
substitutions) EMS
Bleomycin small
deletions UV mostly point mutations but also
large deletions Ionizing radiation (X-,
gamma-rays) large deletions, rearrangements Dosa
ge kill 90 Expression period dilute out
WT molecules (pre-existing protein. mRNA)
Metabolic cooperation WT toxic product can be
transferred.. Dominant vs. recessive mutations
Dom rare (subtle) but expressed, Recessives
easy but masked. Mutagen specificity (a
particular base or base combo). Mutational
spectra (hot and cold spots). Strand
specificity transcribed strand is often
preferentially repaired.
7
- Categories of cell mutants
- Exploitable metabolic pathways
- Purine and pyrimidine biosynthesis auxotrophs
- (auxotrophs require a nutrient in the medium that
the WT doesnt) - Auxotrophs BUdR (BrdU) Kao and Puck. Kill
growing cells. General method. - Analogous to penicillin selection in
prokaryotes. - Many auxotrophs in amino acid, nucleotide
biosynthetic pathways isolated - 2. Drug resistance see sheet
- A. Mutant lacks toxifying enzyme
- e.g., HPRT (TGR), APRT (DAPR, 8-azaAR), TK
(BrdUR) - B. Enzyme target becomes a better discriminator
- (ouabain NaK ATPase pump a-amainitin
RNA Pol II) - C. Permeation changes influx blocked or efflux
increased. (MDR, P-glycoprotein) - D. Improved de-toxification via chelation,
covalent modification, - or overproduction of target (dhfr
MTX-resistance)
8
(No Transcript)
9
(No Transcript)
10
- 3. Temperature-sensitive mutants cell cycle
mutants. - Tritiated amino acid suicide (aa-tRNA
synthetases) - 4. Antibodies. Lysis with complement. Targets
cell surface constituents mostly (e.g., MHC) - 5. Visual inspection at colony level
- A. Sib selection (G6PD)
- B. Replica plating (LDH)
- C. Secreted product (Iganti-Ig IP)
- FACS fluorescence-activated cell sorter.
- 1-D and 2-D fluorescence displays (cell
surface Ag) - Brute force
- (clonal biochemical analysis, e.g.,
electrophoretic variants (e.g., Ig, isozymes) - Direct genotype analysis (rare) (DNA isolation
(via PCR and SSCP, single strand conformational
polymorphism electrophoresis. Or DGGE denaturing
gradient gel electrophoresis. - MHC major histocompatability locus or proteins
G6PD glucose-6-phosphate dehydrogenase LCH
lactate dehydrogenase Ig immunoglobulin
11
Cell fusion (for gene juxtaposition, mapping,
protein trafficking, ) Fusogenic agents PEG,
Sendai virus (syncytia promoting, as
HIV). Heterokaryons (2 nuclei), no cell
reproduction (limited times). (e.g., membrane
fluidity, nuclear shuttling, gene activation
(myoblasts) Hybrids (nuclei fuse, cells
reproduce). Small of heterokaryons. Complement
ation (e.g., auxotrophs with same
requirement) Dominance vs. recessiveness. Chromos
ome loss from hybrids ? Mapping chromosome
assignment. Synteny. Radiation hybrids linkage
analysis (sub-chromosomal regional assignments).
PEG polyethylene glycol, (1000 to 6000 MW)
12
Cell fusion
Hprt, TK-
Parental cells
Hprt-, TK
HAT-
HAT-
PEG (polyethylene glycol, mw 6000 Sendai virus,
inactivated
Cell fusion
Heterokaryon (or, alternatively, homokaryon)
HAT medium
Hprt-, TK, Hprt TK-
HAT
Cell cycle, Nuclear fusion, Mitosis, survival
Hprt-, TK, Hprt TK-
membrane dynamics (lateral diffusion
Edidin), shuttling proteins (hnRNP A1
Dreyfuss), gene regulation (turn on myogenesis
Blau)
Hybrid cell
gene mapping (synteny Ruddle) gene regulation
(extinction Weiss) Complementation (pyrimidine
path Patterson)
13
Complementation analysis
Parental cells
Parental cells
gly-
gly-
gly-
gly-
Cell fusion
Cell fusion
glyA- glyA-
glyA- glyB-
Hybrid cell
Hybrid cell
Glycine-free Medium No growth, no
complementation, ?same gene (named glyA)
Glycine-free Medium Yes, growth, Yes,
complementation, ?different genes genes (named
glyA and glyB)
14
Mapping genes to chromosomes
Hybrid cell
Reduced hybrid
Spontaneous chromosome loss (human
preferentially)
Hprt-, TK, Hprt TK-
Hprt-, TK, Hprt TK-
Correlate identified chromosome loss With loss of
phenotypic trait (isozyme, DNA sequence, etc.)
15
Transfection agents DEAE-dextran (toxic, OK for
transient) CaPO4 (co-precipitate) Electroporation
(naked DNA, high quick voltage ? transient
holes) Lipofection (multilamellar
liposomes) Polybrene (detergent?) Ballistic
(DNA-coated gold particles) Must traverse
cytoplasm. Much engulfed in lysosomes.
Inhibition of lysosomal function often helps
(chloroquin) Pechelosome 2000 KB
co-integration of high MW DNA. Separate plasmids
-gt same site (co-integration). Separate
transfections -gt separate locations Random or
semi-random (many) integration sites (unless
targeted) Low but real homologous recombination
rate History mammalian cell transfection
developed for practical use at Columbia (PS
Wigler Axel and Silverstein)
16
Mike Wigler
Richard Axel
Saul Silverstein
History discovered for practical use at Columbia
(PS Wigler Axel and Silverstein)
17
Transient transfection vs.
permanent cloned genes Unintegrated DNA
chromosomally
integrated Unnatural?
position effects
? Super-physiological expression (so
average many) levels (per transfected cell)
? Transient -gt 10-50 transfection efficiency
(stain) Permanents more like 0.001 per µg DNA
per cell (high). i.e., 106 -gt 1000 colonies
could be much less for certain types of cells
18
One the most dramatic first applications of gene
transfection from total DNA Transfer of the
growth-transformed phenotype ability to grow in
multilayers or in suspension in soft agar
(Weinberg, Wigler) DNA from tumor transfected
into growth controlled mouse 3T3 cells. Look
for foci (focus). Make a library from
growth-transformed transfectant. Screen for human
Alu repeat. Verify cloned DNA yields high
frequency of focus-forming transfectants. Isolate
cDNA by hybridization. Sequence. Identify gene
a dominant oncogene. Ras, a signaling protein
in transducing pathway for sensing growth factors
19
Recombination gene targeting Mitotic
recombination between homologous chromosomes
relation to cancer through the loss of tumor
suppressor genes LOH loss of homozygosity WT
/ ? mutation ? /- (WT phenotype) ? (LOH
via homologous recombination in G2 or chromosome
loss and duplication)? -/- (mutant phenotype
revealed) Recombination of transfecting
genes homologous vs. non-homologous
recombination. Gene conversion vs. reciprocal
recombination. Recombination between tandem
inserts (higher freq).
20
Gene knockouts via homologous recombination.
ES cells and transgenic mice. Selection for
homologous recombinants via the loss of HSV TK
genes (Capecchi) tk homol. region YFG
homol. region tk (YFG your favorite
gene) Allele replacements in cultured cell lines
(e.g., APRT). Most work in ES cells ? mice ?
homozygosis via F1 breeding Little work in
cultured lines Myc double sequenctial K.O.
viable, sick (J. Sedivy) Splicing factor (ASF)
double K.O. in chick DT40 lymphoid cells (high
rate of homologous recombination (J. Manley)
Would be lethal, but cover with inducible human
ASF gene (tet-off) Then add tet to analyze
effects of gene product removal
21
Double knockout of the ASF gene, a vital gene, by
homologous recombination
Chicken DT40 cells
ASF-
neo
Tet-off promoter
pur
neo
ASF-
neo
tet
pur
ASF-
pur
X
Cell dies without ASF
cell viable
Wang, Takagaki, and Manley, Targeted disruption
of an essential vertebrate gene ASF/SF2 is
required for cell viability. Genes Dev. 1996 Oct
1510(20)2588-99.
22
Gene amplification Historically Methotrexate
resistance (Littlefield) High dihydrofolate
reductase (DHFR) enzyme activity, protein,
protein synthetic rate, translatable mRNA.
(Schimke) mRNA level, DNA level. Homogeneously
staining, expanded chromosomal regions (HSRs)
Biedler Nunberg HSR dhfr genes. Double
minute chromosomes. Amplicons. Big (300 KB). Can
shrink, migrate.
23
Tritium grains from hybridized cDNA
24
Gene amplification
Homogeneously staining region FISH, here
25
Original locus?
HSR ? dmin upon DS break induced by a homing
endonuclease (I-SceI).
HSR homogeneously staining region Dmin double
minute chromosomes
Arnaud Coquelle, Lorène Rozier, Bernard
Dutrillaux and Michelle Debatisse ONCOGENE, 31
October 2002, Volume 21, Number 50, Pages
7671-7679 Induction of multiple double-strand
breaks within an hsr by meganucleaseI-SceI
expression or fragile site activation leads to
formation of double minutes and other chromosomal
rearrangements
26
Ampification models over-replication, unequal
sister chromatid exchange, breakage and fusion
(Tanaka paper). Map dhfr amplicons (Schimke,
Hamlin) 300 kb , but wide range Gene
amplification is rare in normal cells (Wahl,
Tslty). p53- allows. In nature rDNA in
oocytes, Drosophila chorion genes. In medicine
chemotherapy resistance (MDR, P-glycoprotein,
efflux pump) cancer (myc, ras) In
biotechnologyhigh level recombinant protein
production in mammalian cells
27
Fred Alt
Geoff Wahl
George Stark
28
Gene amplification for high level production in
CHO dhfr- cells.
29
Reduction of folate to tetrahydrofolate
30
(No Transcript)
31
Biosynthesis of glycine
32
Biosynthesis of TMP
33
Biosynthesis of purine nucleotides
34
DHFR- cells require G,H,T
35
A different major system for high level Mab
production NS0 cells Mouse myeloma cells, high
IgG producers ? IgG variants NS0 No endogenous
IgG, but cell is a natural IgG secretor. Lack
glutamine synthetase (GS) glutamate NH3
ATP ? glutamine ADP Pi Vector MAb genes
driven by strong promoters (H-chain, L-chain)
GS cDNA gene (Bebbington) Select on
glutamine-free medium Inhibit GS with methionine
sulfoximine (gln analog) Select for GS
overproducers ---gt--gt (amplification of the GS
cDNA gene and linked Mab genes) Proprietary
(Lonza Biologics)
36
Some other amplifiable genes
37
Transfection strategies
- YFG (Your Favorite Gene) linked to a dhfr
minigene on a single plasmid - A. Insures co-integration
- B. Insures co-amplification
- YFG and dhfr on separate plasmids
- A. Allows a high ratio of YFG to dhfr to start
38
Linked amp
CHO cells
39
Co-amp1
40
Co-amp3
(with or without pre-ligation)
41
kaufman
42
Co-amp2
43
Co-amp4
44
Amplification protocol