Chapter 14: Migration - PowerPoint PPT Presentation
1 / 29
Title:
Chapter 14: Migration
Description:
Different evolutionary forces on populations = different allele frequencies ... population is divided into many sub-populations, like islands in an archipelago. ... – PowerPoint PPT presentation
Number of Views:54
Avg rating:3.0/5.0
Title: Chapter 14: Migration
1
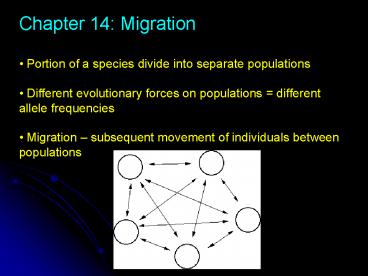
- Chapter 14 Migration
- Portion of a species divide into separate
populations - Different evolutionary forces on populations
different allele frequencies - Migration subsequent movement of individuals
between populations
2
- 14.1 Mating systems
- Random mating
- (choice of mates independent of genotype and
phenotype) - Inbreeding
- (Mating between relatives)
- Preferential mating
- (non-random, based on phenotype and genotype)
- - Assortative mating
- (Same phenotype attracts e.g. human choice
based on intelligence) (Mm) - - Disassortative mating
- (Self-sterility systems in plants)
3
- 14.2 Estimating migration
- Migration is the movement of individuals from
one breeding population to another. - Assume that random individuals are involved.
- After migration Pop 1 increase in size by
proportion m, which comes from Pop 2. - n2 number of migrants, n1 native pop size
- If the frequency of a given gene is q2 among
immigrants and q1 among the natives, the new
frequency will be - qm m(q2 - q1) q1 and m n2 / (n1 n2)
4
Example For given Pop 1, n 8,000 2,000
animals from Pop 2 emigrates to Pop 1 q1 was
0.2 q2 0.6 m n2 / (n1 n2) 2,000 /
(8,000 2,000) 0.2 The new frequency of q
after migration (qm) is qm m(q2 - q1)
q1 0.2 (0.6 0.2) 0.2 0.28 Change in
allelic frequency ?q 0.28 - 0.2 0.08
5
- 14.3 The island model of migration
- In this model, the overall population is divided
into many sub-populations, like islands in an
archipelago.
6
- 14.3 The island model of migration
- In this model, the overall population is divided
into many sub-populations, like islands in an
archipelago. - Each sub-population is so large that random
genetic drift is not possible. - Consider two alleles, A and a ( p and q)
- The amount of migration is m
- The generation is t
- P in each population is constant
- (because of infinite size)
- pt p (po p)(1 m)t
7
Suppose there are 2 populations, with initial
frequencies of A 0.2 and A 0.8 respectively
and m 0.1 What is the frequency of A in the 2
populations after 10 generations? With movement
of migrants in both direction, the frequency of A
in migrants (0.2 0.8) / 2 0.5 For Pop
that started with A 0.2 P10 0.5 (0.2
0.5)(1 - 0.1)10 0.395 For Pop that started
with A 0.8 P10 0.5 (0.8 0.5)(1 - 0.1)10
0.605
8
The change in allele frequency with time, in five
sub-populations exchanging migrants at a rate of
m 0.1 per generation
Because migration rates are much greater than
mutation, changes in allele frequencies occur
much faster with migration.
9
14.4 One-way migration When migration occurs
from one population onto another, without an
equal migration in the reverse direction.
10
- 14.5 Isolate breaking and Wahlunds principle
- Isolate breaking fusion of formerly isolated
sub-populations, by migration. - Fusion reduces the frequency of homozygotes
- Wahlunds principle
11
- 14.6 How migration limits genetic divergence
- A relatively low level of migration between
populations can prevent significant divergence
between the populations. - Migration can be expressed as Nm, which denotes
the number of migrants per generation. - Nm is derived from F
- When F 1, Nm 0
12
- 14.7 Patterns of migration (actual patterns)
- Migration in real populations is more complex
than is assumed in the island model of migration. - In nature, migrants come mostly from nearby
populations (as opposed to several random
populations) - Humans migration rates depend on age, sex,
status, population density. - Managed wildlife populations ?
13
- 14.8 Genetic distance
- A coefficient to express genetic differentiation
among populations. - Several varieties exist, with the most common
measure - Neis genetic distance or D
- D -ln(I)
- Where I genetic identity the correlation of
allele frequencies between 2 populations.
- Possible values for D can vary from 0 (no
difference) to 1 (no similarity).
14
Example For 3 populations and 1 locus with 3
alleles
Genetic distance between populations 1 and
2 For Pop1 ?pix2 (0.023)2 (0.977)2
(0.0)2 0.955 For Pop2 ?piy2 (0.019)2
(0.885)2 (0.096)2 0.793 And ?(pixpiy)
(0.023x0.019) (0.977x0.885) (0.0x0.096)
0.865
15
Genetic distance between populations 1 and
2 For Pop1 ?pix2 (0.023)2 (0.977)2
(0.0)2 0.955 For Pop2 ?piy2 (0.019)2
(0.885)2 (0.096)2 0.793 And ?(pixpiy)
(0.023x0.019) (0.977x0.885) (0.0x0.096)
0.865
IN ?(pixpiy) / v (?pix2 )(?piy2) 0.865
/ v (0.955 x 0.793) 0.994 And genetic
distance -ln(IN) -ln(0.994) 0.006
16
Chapter 15. Mutation A sudden (and heritable)
change in the genetic material
17
Mutation
- With each generation gene pool is shuffled
produce new genotypes (combinations)
- Large number of possible combinations only a
fraction in living members of the population
(for example 5 loci 3 alleles give 7,776
possible genotypes)
- Enormous genetic reserve - produce new
genotypic combinations continuously
- Assortment and recombination do not produce new
alleles
18
Mutation create new alleles
Random process, without regard for any benefits
or disadvantages to the organism
19
- 15.1 Number of alleles maintained in populations
- An average protein contains 300 amino acids
900 nucleotides - The number of possible alleles 10542
- We can therefore assume that every time a
mutation occurs, it is a new allele that does not
already exist in the population. - the Infinite allele model of mutation.
20
- 15.2 The neutrality hypothesis
- The neutrality hypothesis predicts that many
mutations have such mild effects, that their
influence on survival and reproduction are
negligible. - The frequencies of such alleles are determined
by forces such as migration and random genetic
drift, instead of selection. - (The evidence for this hypothesis comes from
early work on allozymes, i.e. protein phenotypes
used as indicators of underlying genotypes).
21
- 15.3 Mutations and behaviour
- Mutations that effect processes in the brain
result in different alleles that cause conditions
such as Huntingtons disease, PKU and
Schizophrenia.
- The mechanism of mutations
- Mutations in egg and sperm cells will be
transmitted unless natural selection intervenes. - Most mutations are not translated into proteins,
because they occur in DNA regions that are not
transcribed (introns). They thus have no visible
effect.
22
A single-base mutation results in the insertion
of a different amino acid into a protein. (
altered function) e.g. ATT TAC
CGC becomes ATT TCC CGC TAC codes for
methionine TCC codes for arginine The effect
of replacing a single amino acid can be anything
from small to lethal.
23
A deletion is usually more damaging than a
substitution, because the entire reading frame of
the DNA triplet code is shifted e.g. TAC AAC
CAT After the loss of C becomes TAA ACC AT-
24
- 15.4 Targeted mutations
- Techniques used during genetic modification of
organisms, by which genes are changed in a
specific way to alter their function. - Knock out deleting key areas of DNA sequence,
preventing the gene from being transcribed. - Newer techniques alter gene expression in more
subtle ways, by inducing underexpression or
overexpression. - In the field of neurogenetics, targeted
mutations are used to elucidate processes such as
learning and memory (in suitable laboratory
animals).
25
Mutation rate
- A gene undergoes mutation to a dominant allele
- 2 out of 100,000 births exhibit mutant phenotype
- Parents are phenotypically normal
- Zygote carries two copies of the gene
- Study 200,000 copies of the gene
- Assume affected births are heterozygous
- Uncovered 2 mutant alleles out of 200,000
26
The mutation rate is 2 / 200,000 1 /
100,000 1 x 10
-5
27
Example
Achondroplasia dominant form of
dwarfism Individuals enlarged skull, short arms
and legs Diagnosed by X-ray at birth Mutation
rate 0.5 x10 Estimate the extent to which
mutation can cause allele frequencies to change
from one generation to the next
- 5
d ? normal alleles D ? allele for achondroplasia
28
- Population 500,000 individuals
- 100 dd
- Initial frequencies d 1.0
- D 0.0
- Each individual contributes 2 gametes
- Gene pool contains 1,000,000 gametes
- Assume 1.4 of every 100,000 d alleles mutates
into a D allele
29
Frequency of allele d (1,000,000 -14)
1,000,000
0.999986
Frequency of allele D ___14___
1,000,000
0.000014
It will take 70,000 generations to reduce the
frequency of allele d from 1.0 to 0.5





![❤️[READ]✔️ Handbook on Migration and Ageing (Elgar Handbooks in Migration) PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10051246.th0.jpg?_=202406081111)
![❤️[READ]✔️ Handbook on Migration and Ageing (Elgar Handbooks in Migration) PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10050658.th0.jpg?_=202406081212)























