NMR of protein - PowerPoint PPT Presentation
1 / 31
Title:
NMR of protein
Description:
In the a-helix the neighbouring HN are 2.8 ngstr m apart ... If 2=Val (V), 5 from a tryptophan (W) and 9 from a glycine (G), we can write that sequence is ... – PowerPoint PPT presentation
Number of Views:96
Avg rating:3.0/5.0
Title: NMR of protein
1
NMR of protein
2
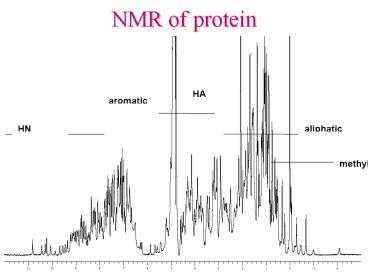
1H NMR spectra of small protein
3
Protein folded and unfolded
Folded
Unfolded
4
2D-COSY
5
2D-COSY NH/CHa expanded region
6
2D-NOESY
7
sequential assignment using NOESY
- In the a-helix the neighbouring HN are 2.8
Ångström apart - In the ß-strand the distance from Ha in residue
(i) to HN in residue (i1) is only 2.2 Ångström.
8
Schematic presentation NOESY spectrum in the NH
region
The diagonal cross peaks are marked as black
signals labeled 1 to 9. The red cross peaks are
sequential NOEs.
If 2Val (V), 5 from a tryptophan (W) and 9 from
a glycine (G), we can write that sequence
is XVXXXWXXG or GXXWXXXVX 983567421 sequence of
signals GAKWSRYVP amino acid sequence
1 ? 2 ? 4 ? 7 ? 6 ? 5 ? 3 ? 8 ? 9
Or reverse
9
Sequential Assignment of Ha in residue (i) to NH
in residue (i1) by NOE
Superimposition of a COSY spectrum with blue
annotated cross peaks (through bond) and NOESY
spectrum with red cross peaks. Sequential NOE
between Ha in residue (i) to HN in residue (i1).
CH1/NH5
6 ? 4 ? 1? 5 ? 7 ? 3 ? 2
CH4/NH1
CH6
NH6
NH4
10
NOE-distance
using distance constraints to calculate a
structure. In the upper figure is shown a linear
strand with beads. The green, red and blue pairs
of beads, respectively have been shown to be
close to each other. The structure below
represents one solution to determining the
structure based on the three pieces of distance
information.
11
Coupling values to setup experiments
140 Hz
H
15 Hz
11 Hz
55 Hz
13C
13C
15N
13C
15N
O
O
H
H
13C
90-100 Hz
30-40 Hz
12
3D-HMQC-COSY
13
HMQC-NOESY
NH 1
2
Ha1
Ca1
2
3
Ha2
Ca2
3
4
Ca3
14
HMQC-NOESY
R. R. Ernst (nobel lecture 92)
15
3D-HMQC-NOESY
16
2D-NOESY vs 3D-HMQC-NOESY
17
NOESY-HMQC
Ha-1
Ha1
NH1
Ha2
NH2
Ha1
Ha3
Ha2
NH3
L.E.Kay, D.Marion, A.Bax, J.Magn.Reson.,84, 72
(89)
18
2D and 3D HMQC-TOCSY
19
3D HCCH-TOCSY
20
3D-NMR Puzzle approach
HNCA
HN(CO)CA
HNCO
21
3D-NMR Puzzle approach
HA(CA)NH
HACACO
HCA(CO)N
22
HNCACB CBCA(CO)NH
23
(No Transcript)
24
Figure 19. Schematic presentation of the combined
spectrum analysis of the three-dimensional HNCACB
and a CBCA(CO)NH spectra. The 15N axis and the
frames in the 15N dimension are coloured blue.
The1H axis and the frames in the 1H dimension are
coloured red. The 13C axis and the frames in the
13C dimension are green. Two planes dN(i),
top-left, and dN(i1), bottom-left, are
highlighted. The cross peaks in HNCACB are and
in CBCA(CO)NH are
25
HNCO 2D and 3D
26
HN(CA)CO and HNCOcomplementary experiments
27
(No Transcript)
28
Example of HNCA and HN(CO)CA
29
Example of sequential assignment
30
2D HN(CO)(CA)
NH
31
4D-NMR