Significance analysis of microarrays (SAM) - PowerPoint PPT Presentation
Title:
Significance analysis of microarrays (SAM)
Description:
Significance analysis of microarrays ... multi-class censored survival one ... Rank the permuted d-values of the genes in ascending order v) Repeat steps iii) ... – PowerPoint PPT presentation
Number of Views:128
Avg rating:3.0/5.0
Title: Significance analysis of microarrays (SAM)
1
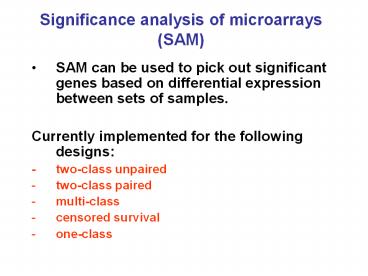
Significance analysis of microarrays (SAM)
- SAM can be used to pick out significant genes
based on differential expression between sets of
samples. - Currently implemented for the following designs
- - two-class unpaired
- two-class paired
- multi-class
- censored survival
- one-class
2
SAM
- SAM gives estimates of the False Discovery Rate
(FDR), which is the proportion of genes likely to
have been wrongly identified by chance as being
significant. - It is a very interactive algorithm allows users
to dynamically change thresholds for significance
(through the tuning parameter delta) after
looking at the distribution of the test
statistic.
3
SAM designs
- Two-class unpaired to pick out genes whose mean
expression level is significantly different
between two groups of samples (analogous to
between subjects t-test). - Two-class paired samples are split into two
groups, and there is a 1-to-1 correspondence
between an sample in group A and one in group B
(analogous to paired t-test).
4
SAM designs
- Multi-class picks up genes whose mean expression
is different across gt 2 groups of samples
(analogous to one-way ANOVA) - Censored survival picks up genes whose
expression levels are correlated with duration of
survival. - One-class picks up genes whose mean expression
across experiments is different from a
user-specified mean.
5
SAM Two-Class Unpaired
- Assign experiments to two groups, e.g., in the
expression matrix - below, assign Experiments 1, 2 and 5 to group A,
and - experiments 3, 4 and 6 to group B.
2. Question Is mean expression level of a gene
in group A significantly different from mean
expression level in group B?
6
SAM Two-Class Unpaired
Permutation tests
- For each gene, compute d-value (analogous to
t-statistic). This is - the observed d-value for that gene.
- ii) Rank the genes in ascending order of their
d-values.
iii) Randomly shuffle the values of the genes
between groups A and B, such that the reshuffled
groups A and B respectively have the same number
of elements as the original groups A and B.
Compute the d-value for each randomized gene
Original grouping
Randomized grouping
7
SAM Two-Class Unpaired
iv) Rank the permuted d-values of the genes in
ascending order
v) Repeat steps iii) and iv) many times, so that
each gene has many randomized d-values
corresponding to its rank from the
observed (unpermuted) d-value. Take the average
of the randomized d-values for each gene. This
is the expected d-value of that gene.
vi) Plot the observed d-values vs. the expected
d-values
8
SAM Two-Class Unpaired
Significant positive genes (i.e., mean
expression of group B gt mean expression of
group A)
Observed d expected d line
The more a gene deviates from the observed
expected line, the more likely it is to be
significant. Any gene beyond the first gene in
the ve or ve direction on the x-axis (including
the first gene), whose observed exceeds the
expected by at least delta, is considered
significant.
Significant negative genes (i.e., mean
expression of group A gt mean expression of group
B)
9
SAM Two-Class Unpaired
- For each permutation of the data, compute the
number of positive and negative significant genes
for a given delta as explained in the previous
slide. The median number of significant genes
from these permutations is the median False
Discovery Rate. - The rationale behind this is, any genes
designated as significant from the randomized
data are being picked up purely by chance (i.e.,
falsely discovered). Therefore, the median
number picked up over many randomizations is a
good estimate of false discovery rate.