CASE7 - PowerPoint PPT Presentation
1 / 10
Title:
CASE7
Description:
A comparison of the genetic maps to the published Vitisvinifera genome revealed most of the markers showed good linear agreement indicating conservation among the ... – PowerPoint PPT presentation
Number of Views:99
Avg rating:3.0/5.0
Title: CASE7
1
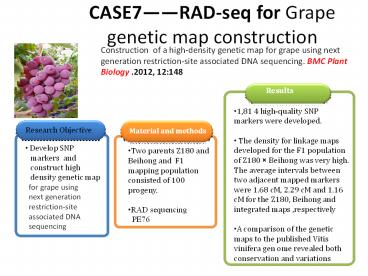
CASE7RAD-seq for Grape genetic map construction
Construction of a high-density genetic map for
grape using next generation restriction-site
associated DNA sequencing. BMC Plant Biology
.2012, 12148
2
An average of 12,840 reads involved in the
polymorphic loci and a 17.0-fold coverage per
cluster per each individual.
3
1,121 markers fell into 19 LGs for Z180 (female),
759 markers for the Beihong (male), and 1,646
markers f or the integrated map.
4
A comparison of the genetic maps to the published
Vitis vinifera genome revealed most of the
markers showed good linear agreement indicating
conservation among the different grape species
and the non-collinearity for some chromosome
regions might indicate some variations among
different grape species during evolution.
5
CASE8RAD-seq assist anthracnose disease
resistance study in Lupinus angustifolius L.
Application of next-generation sequencing for
rapid marker development in molecular plant
breeding a case study on anthracnose disease
resistance in Lupinus angustifolius L. BMC
Genomics 2012, 13318
- discovered 8207 SNP markers.
- 38 markers linked to the disease resistance gene
Lanr1. - Five randomly selected markers were converted
into cost-effective,simple PCR-based markers. - Linkage analysis confirmed that randomly
selected five markers were linked to the R gene.
- Twenty informative plants from a cross of RxS
(disease resistant x susceptible) in lupin were
subjected to RAD single-end sequencing by
multiplex identifiers. - Strategy
- PE91
- RAD
The objectives of this research were to examine
the utility of RAD sequencing, applied as DNA
fingerprinting, for rapid marker development for
MAS in plant breeding, and to develop molecular
markers more closely linked to the disease.
6
A flow diagram illustrating the marker
development procedures in this study
7
Two of the newly developed markers, AnSeq3
and AnSeq4, were flanking the R gene at a genetic
distance of 0.9 cM
8
CASE9 Double enzyme digestion
Genotyping-by-sequencing
- map over 34,000 SNPs and 240,000 tags onto the
Oregon Wolfe Barley reference map - 20,000 SNPs and 367,000 tags on the Synthetic
SynOpDH wheat reference map - D-genome markers was lower than either the A or
B-genomes in Wheat - assembled 1,485 of the SNP markers into a genetic
linkage map of 21 linkage groups.
Material BarleyOWB WheatSynOpDH EnzymePstI
(CTGCAG)MspI (CCGG) Sequencing Illumina GAII/
Illumina HiSeq 2000 PE 64bp
Background
GBS uses restriction enzymes for targeted
complexity reduction followed by multiplex
sequencing to produce high-quality polymorphism
data at a relatively low per sample cost.
9
Figure 1. Adapter Design, PCR amplification of
fragments.
Figure 2. Histogram of number of markers in the
three wheat genomes for DArT and GBS SNP genetic
maps
10
Figure 3. Distribution of GBS SNP markers in the
Oregon Wolfe Barley (OWB) bin map.