Objectives of DNA recombination - PowerPoint PPT Presentation
1 / 114
Title:
Objectives of DNA recombination
Description:
Homologous recombination of E. coli Identification of genes involved in ... isolation of mutants affecting recombination in recB recC sbcB or recB ... – PowerPoint PPT presentation
Number of Views:315
Avg rating:3.0/5.0
Title: Objectives of DNA recombination
1
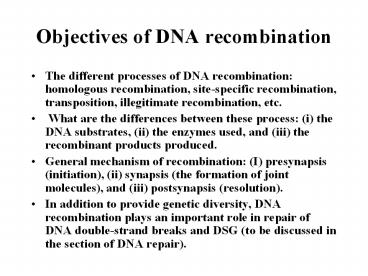
Objectives of DNA recombination
- The different processes of DNA recombination
homologous recombination, site-specific
recombination, transposition, illegitimate
recombination, etc. - What are the differences between these process
(i) the DNA substrates, (ii) the enzymes used,
and (iii) the recombinant products produced. - General mechanism of recombination (I)
presynapsis (initiation), (ii) synapsis (the
formation of joint molecules), and (iii)
postsynapsis (resolution). - In addition to provide genetic diversity, DNA
recombination plays an important role in repair
of DNA double-strand breaks and DSG (to be
discussed in the section of DNA repair).
2
Examples of recombination
3
Homologous recombination
- Refer to recombination between homologous DNA
sequence in the same or different DNA molecules. - The enzymes involved in this process can catalyze
recombination between any pair of homologous
sequences, as long as the size of homologous
sequence is longer than 45 nt or longer. No
particular sequence is required. - Models of homologous recombination.
- Homologous recombination of E. coli.
- Meiotic recombination.
4
The Holliday model of recombination
5
(No Transcript)
6
Homologous recombination of E. coli
- Identification of genes involved in
recombination (i) isolation of mutants affecting
recombination in wild-type cells (eg., recA,
recB, recC etc.), (ii) the recombinational
deficiency in recBC cells may be suppressed by
sbcA or sbcB mutations. The sbcB gene encodes for
a 3 to 5 ss-DNA exonuclease, while the sbcA
mutation activate the expression of recE which
encodes for 5 to 3 exonuclease. (iii) isolation
of mutants affecting recombination in recB recC
sbcB or recB recC sbcA cells (eg., recF, recO,
recR, recQ, recJ etc.) - The biochemical functions of rec genes.
7
Homologous recombination is catalyzed by enzymes
- The most well characterized recombination enzymes
are derived from studies with E. coli cells. - Presynapsis helicase and/or nuclease to generate
single-strand DNA with 3-OH end (RecBCD) which
may be coated by RecA and Ssb. - Synapsis joint molecule formation to generate
Holliday juncture (RecA). - Postsynapsis branch migration and resolution of
Holliday juncture (RuvABC).
8
RecBCD
- A multifunctional protein that consists of three
polypeptides RecB (133 kDa), RecC (129 kDa) and
RecD (67 kDa). - Contain nuclease (exonuclease and Chi-specific
endonuclease) and helicase activity.
9
Chi-specific nicking by RecBCD
5-GCTGGTGG-3
Fig. 22.7
10
Helicase and nuclease activities of the RecBCD
11
The Bacterial RecBCD System Is Stimulated by chi
Sequences
FIGURE 15.17 RecBCD unwinding and cleavage
12
The RecBCD pathway of recombination
13
RecA binds selectively to single-stranded DNA
Fig. 22.4
14
RecA forms nucleoprotein filament on
single-strand DNA
15
Fig. 22.5
16
Paranemic joining of two DNA (in contrast to
plectonemic)
Fig. 22.6
17
Strand-Transfer Proteins Catalyze Single-Strand
Assimilation
- RecA forms filaments with single-stranded DNA and
catalyzes the assimilation of single-stranded
DNA to displace its counterpart in a DNA duplex.
FIGURE 15.18 RecA strand invasion
18
RuvABC
- RuvA (22 kDa) binds a Holliday junction with high
affinity, and together with RuvB (37 kDa)
promotes ATP-dependent branch migration of the
junctions leading to the formation of
heteroduplex DNA. - RuvC (19 kDa) resolves Holliday juncture into
recombinant products.
19
Fig. 22.9
20
Fig. 22.10
21
(No Transcript)
22
(No Transcript)
23
Fig. 22.14
24
Fig. 22.15
25
Fig. 22.17
26
(No Transcript)
27
(No Transcript)
28
Homologous Recombination Occurs between Synapsed
Chromosomes in Meiosis
- Chromosomes must synapse (pair) in order for
chiasmata to form where crossing-over occurs. - The stages of meiosis can be correlated with the
molecular events at the DNA level.
FIGURE 03 Recombination occurs at specific
stages of meiosis
29
Fig. 15.13
30
(No Transcript)
31
Fig. 15.15
32
The Synaptonemal Complex Forms after
Double-Strand Breaks
- Double-strand breaks that initiate recombination
occur before the synaptonemal complex forms. - If recombination is blocked, the synaptonemal
complex cannot form. - Meiotic recombination involves two phases one
that results in gene conversion without
crossover, and one that results in crossover
products.
33
Fig. 22.18
34
Fig. 22.19
35
Fig. 22.20
36
Fig. 22.21
37
(No Transcript)
38
Fig. 22.24
39
Gene conversion the phenomenon that abnormal
ratios of a pair of parental alleles is detected
in the meiotic products.
40
Fig. 22.25
41
Fig. 22.26
42
Site-specific Recombination Bacteriophage lambda
integration in E. coli
43
Fig. 15.28
44
A site-specific recombination reaction (eg.
catalyzed by Int of bacteriophage lambda)
45
Fig. 15.31
46
(No Transcript)
47
(No Transcript)
48
(No Transcript)
49
(No Transcript)
50
Recombination Pathways Adapted for Experimental
Systems
FIGURE 15.38 Cre/lox system for gene knockouts
Adapted from H. Lodish, et al. Molecular Cell
Biology, Fifth edition. W. H. Freeman Company,
2003.
51
Fig. 23.21
52
Fig. 23.12
53
Fig. 23.13
54
Fig. 23.14
55
Fig. 23.15
56
Fig. 23.16
57
Fig. 23.17
58
Transposition
- Transposition is mediated by transposable
elements, or transposons. - Transposons of bacteria IS (insertion sequences)
contains only sequences required for
transposition and proteins (transposases) that
promote the process. Complex transposons contain
genes in addition to those needed for
transposition. - Transposition is characterized by duplication of
direct repeats (5-9 bps in most cases) at target
site. - Transposition, in some instances, may be mediated
through a RNA intermediate (retrotransposons and
non-LTR retrotransposons).
59
Duplication of the DNA sequence at a target site
when a transposon is inserted
60
Fig. 23.1
61
Fig. 23.2
62
Fig. 17.3
63
(No Transcript)
64
Fig. 23.3
65
Fig. 23.4
66
Fig. 23.5
67
Replicative transposition is meidated by a
cointegrate intermediate.
Fig. 23.6
68
Fig. 23.7
69
Eukaryotic transposons
- DNA transposons (i) Ds and Ac of maize, (ii)
Drosophila P elements. - Retrotransposons (i) LTR retrotransposons (Ty
element of yeast and copia of Drosophila). (ii)
non-LTR retrotransposons (LINES, Alu, group II
introns).
70
Ds and Ac of maize
Fig. 23.8
71
Fig. 23.9
72
Fig. 23.10
73
Hybrid Dysgenesis
Fig. 17.20
F
74
Fig. 17.21
75
Fig. 17.22
76
Fig.23.19
77
Fig. 23.18
78
Fig. 23.20
79
Fig. 23.21
80
Fig. 23.22
81
Fig. 23.23
82
(No Transcript)
83
Fig. 23.24
84
Nonviral transposons LINES
Fig. 23.25
85
Fig. 23.26
86
Fig. 23.27
87
Fig. 23.28
88
Group II introns Retrohoming
89
DNA Repair
- DNA damage may arise (i) spontaneously, (ii)
environmental exposure to mutagens, or (iii)
cellular metabolism. - DNA damage may be classified as (I) strand
breaks, (ii) base loss (AP site), (iii) base
damages, (iv) adducts, (v) cross-links, (vi)
sugar damages, (vii) DNA-protein cross links. - DNA damage, if not repaired, may affect
replication and transcription, leading to
mutation or cell death.
90
Fig. 20.27
91
Fig. 20.28
92
Fig. 20.29
93
(No Transcript)
94
Methylataion and Mismatch Repair
95
Model for Mismatch Repair
96
(No Transcript)
97
(No Transcript)
98
Base-Excision Repair
99
FIGURE 16.12 Uracil is removed from DNA
FIGURE 16.13 Glycosylases remove bases
100
16.5 Base Excision Repair Systems Require
Glycosylases
FIGURE 16.14 Base removal triggers excision
repair
101
Nucleotide-Excision Repair in E. coli and Humans
102
(No Transcript)
103
Alkylation of DNA by alkylating agents
104
Direct Repair Photoreactivation by photolyase
105
O6-methyl G, if not repaired, may produce a
mutation
106
Direct Repair Reversal of O6 methyl G to G by
methyltransferase
107
Direct repair of alkylated bases by AlkB.
Direct re
108
Effect of DNA damage on replication (i) coding
lesions wont interfere with replication but may
produce mutation, (ii) non-coding lesions will
interfere with replication and may lead to
formation of daughter-strand gaps (DSG) or
double-strand breaks (DSB).
DSG and DSB may be repaired by recombination
process, to be discussed in the following section.
109
Models for recombinational DNA repair
110
Fig. 20.40
111
(No Transcript)
112
Fig. 20.38
Model for nonhomologous end-joining
113
Figure 16.25 NHEJ requires several reactions.
114
Fig. 20.41