Problems with dnds test - PowerPoint PPT Presentation
1 / 31
Title:
Problems with dnds test
Description:
Advantageous & deleterious. dn ds. dn ds. Combining Data. between. within ... are deleterious or neutral. McDonald & Kreitman (1991) Nature. McDonald-Kreitman Test ... – PowerPoint PPT presentation
Number of Views:72
Avg rating:3.0/5.0
Title: Problems with dnds test
1
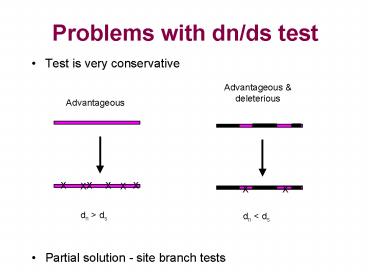
Problems with dn/ds test
- Test is very conservative
- Partial solution - site branch tests
Advantageous deleterious
Advantageous
X
X
X
X
X
X
X
X
dn gt ds
dn lt ds
2
Combining Data
between
within
3
Human1 CCC GCA GAG TTA CTA ATC GAA Human2 CCG GCA
GAG TTA CTA ATC GAA Human3 CCC GCA AAG TTA CTA
ATC GAA Human4 CCC GCA AAG TTA CTA ATC GAA Chimp
CCC GCC GAG TTA GTA ATT GAA
4
McDonald-Kreitman Test
Assume - synonymous mutations are neutral -
non-synonymous / amino acid mutations
are deleterious or neutral
McDonald Kreitman (1991) Nature
5
McDonald-Kreitman Test
6
Neutral v advantageous
Frequency
Time ?
7
McDonald-Kreitman Test
8
McDonald-Kreitman Test
9
Example jingwei
New gene found in D.yakuba and D.teisseri
Long Langley (1994) Nature
10
Jingwei - dn / ds
D.teisseri
D.yakuba
11
jingwei
D.teisseri
D.yakuba
12
How Much Adaptive Evolution ?
13
jingwei
? 95
D.teisseri
D.yakuba
14
D.simulans D.yakuba
600,000 aa differences
15
Genomic Estimates - Drosophila
115 genes - multiple alleles in D.simulans -
one allele in D.yakuba
? 49
Welch (2006) Genetics
16
D.simulans D.yakuba
600,000 aa differences 41 adaptive 246,000
adaptive 1 every 49 years
17
Enteric Bacteria
E.coli
S.enterica
gt55 genes shared
gt37,000 aa diffs
18
Dataset
- Charlesworth and EW (2006) MBE
- 6 complete E.coli genomes
- 6 complete S.enterica genomes
- 410 genes present in all strains
19
Data
? -0.65
20
Deleterious Mutations
21
Slightly Deleterious Mutations
22
Deleterious
Frequency
Time ?
23
Slightly Deleterious Mutations
S.enterica
Non-synonymous
Synonymous
24
Data
? -0.65
25
Deleterious
Frequency
Time ?
26
Deleterious
Frequency
Time ?
27
Deleterious
Frequency
Time ?
28
Adaptive Evolution
S.enterica
? 0.34 (0.14, 0.50)
29
Adaptive Evolution
E.coli
? 0.58 (0.45, 0.68)
30
Enteric Bacteria
E.coli
S.enterica
55 genes shared
37,000 aa diffs
21,000 adaptive aa diffs
1 every 8000 years
31
Further reading
- Eyre-Walker (2006) TREE