Differential Gene Expression - PowerPoint PPT Presentation
1 / 32
Title:
Differential Gene Expression
Description:
maskin: protein in Xenopus that blocks mRNA translation by binding to. the 5' and 3' UTRs ... Vg1 in Xenopus eggs is another-- localized to the vegetal (yolk) half ... – PowerPoint PPT presentation
Number of Views:37
Avg rating:3.0/5.0
Title: Differential Gene Expression
1
Differential Gene Expression
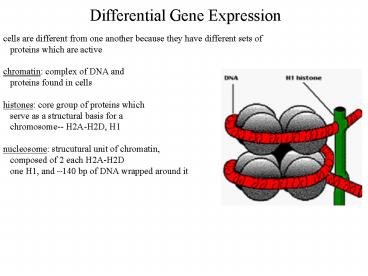
cells are different from one another because they
have different sets of proteins which are
active chromatin complex of DNA and proteins
found in cells histones core group of proteins
which serve as a structural basis for a
chromosome-- H2A-H2D, H1 nucleosome strucutural
unit of chromatin, composed of 2 each H2A-H2D
one H1, and 140 bp of DNA wrapped around it
2
Differential Gene Expression
condensed DNA (ie. wrapped up with histones) is
inactive-- RNA polymerases can't access the
DNA very effectively acetylation and methylation
of histones regulate their ability to form
nucelosomes (acetylation disruption,
methylationstabilize) and therefore their
ability to make mRNA and, indirectly,
protein usually affects a group of genes--
depends upon blocking promoters
3
Differential Gene Expression
TATA box
mRNA transcription
enhancers
transcriptional start site
silencers
promoter
upstream elements
downstream sequence
promoter region of DNA that binds the RNA
polymerase complex to initiate
transcription TATA box sequence of DNA usually
located within 30bp of where transcription
will start that physically interacts with RNA
polymeraseII RNA polymerase II makes all
mRNAs, but not tRNA or rRNA
4
Differential Gene Expression
TATA box
mRNA transcription
enhancers
transcriptional start site
silencers
promoter
upstream elements
downstream sequence
transcription factors proteins which bind to
DNA sequences to regulate the process of
transcription basal transcription factors
minimal set of transcription factors required
regulated transcription factors transcription
factors that change their expression-- makes
cells different from one another
5
Differential Gene Expression
TATA box
mRNA transcription
enhancers
transcriptional start site
silencers
promoter
upstream elements
downstream sequence
silencers DNA sequences which bind repressors
that reduce transcription enhancers DNA which
binds activators that increase transcription upst
ream elements DNA sequences that are 5' of
transcription downstream seqeunce DNA sequence
3' of promoter which is transcribed
6
Differential Gene Expression
RNA polymerase II
mRNA transcription
activators
transcriptional start site
repressors
basal transcription factors
upstream elements
downstream sequence
activator protein which binds to enhancers to
increase transcription repressor protein which
binds to silencers to decrease transcription acti
vators and repressors can act far from the
transcriptional start site most function 5' of
promoters (upstream) but not a requirement
7
Assembly of the Polymerase Complex
enhancers bound by activators work with
coactivators to increase transcription Silencer
elements would be bound by repressors and work
with corepressors to block TFIID or other
general transcription factor TAFs and mediator
complex work to further regulate
binding Looping of DNA is vital several dozen
proteins work together for RNA pol II to start
8
Generalizations about Regulatory DNA elements
most genes require enhancers to be
transcribed enhancers are the most common
temporal and spatial regulator of mRNA multiple
enhancers at several sites work together to
regulate expression distal transcription factors
interact with basal factors at the promoter
precise mechanisms are not well
understood enhancers are combinatorial--
different pieces can be mixed and
matched enhancers are modular-- different
enhancers are active at different places or
times enhancers function in 2 ways-- either
modulate chromatin or pol II binding same rules
for enhancers apply for silencers
9
Transcription Factors
Transcription factors have 3 major domains 1)
DNA binding domains- must interact with
particular sequences of DNA
leucine zipper
helix- loop- helix
10
Transcription Factors
2) trans-activating domain- activates or
suppresses the promoter of interest note that
this is similar to 3-- this domain is generally
going to function somewhat at a distance but
work directly on the basal factors
3) protein-protein interaction domain-- binds to
proteins that will change the function of the
transcription factor may interact with basal
transcription factors, but does not have to
11
trans-Activating domains
trans-activating domains need to regulate a
promoter one of the more common mechanisms is
to affect nucleosomes histone acetyltransferase
enzyme which transfers acetyl groups to histones
and causes nucleosomes to destabilize (unwind)
usually associated with activators (proteins
that bind to enhancers) histone deacetylase
enzyme which removes acetyl groups from histones
and increases nucleosome formation usually
associated with repressors (proteins that bind
silencers) different factors recruit histone
modifying proteins to particular locations
12
trans-Activating domains
acetylation destabilizes nucleosomes histone
methyltransferase enzyme which transfers a
methyl group to a histone protein-- usually to
the same amino acid that can receive an acetyl
group methylation stabilizes nucleosomes by
preventing histone acetylation
dynamic equilibrium balanced processes leading
to a stable situation
13
DNA Methylation and Transcriptional Control
histones are not the only part of chromatin that
gets methylated cytosines in DNA are also
methylated-- only occurs on CG sequences!
5-methylcytosine stabilizes nucleosomes (same as
histone methylation) not all organisms-- fruit
flies and C elegans don't methylate their DNA
DNA becomes methylated during the process of
differentiation when cells divide, one strand
of DNA goes to each daughter when one strand
is methylated, the other strand also gets
methylated, hence the requirement for CG
sequences helps the cell to 'remember' which
genes are active
5' 3' G-Cm mC-G 3' 5'
5' 3' G-C mC-G 3' 5'
5' 3' G-Cm C-G 3'
5'
cell division
14
DNA Methylation and Transcriptional Control
DNA methylation is important for several major
reasons for cloning, cells must be stopped in G2
phase of their cell cycle, right after DNA
replication-- ie. newly replicated DNA is
unmethylated blastomeres have less methylated
DNA than more differentiated cells genomic
imprinting genes inherited from either the
mother or the father are more likely to be
expressed than from the other parent 30 known
inactive chromosome is significantly more
methylated than active one dosage compensation
inactivation of 1 X-chromosome in females
directed by a particular mRNA Xist directs one
chromosome to become methylated-- the other
chromosome locally inhibits Xist leads to the
heritable barr body
15
Methods for Studying Regulatory DNA
reporter gene protein which, when examined
properly, shows where it is expressed and at
what level b-galactosidase is an enzyme
responsible for sugar metabolism in bacteria
converts a colorless NBT (nitro blue tetrazolium
salt) to a blue precipitate-- blue color is
visible even in (small) whole embryos GFP green
fluorescent protein fluoresces green under
blue light small, doesn't affect most
proteins visible in living animals!!!
originally isolated from jellyfish now
available in various colors multiple colors
allow several proteins to be followed at once
16
Methods for Studying Regulatory DNA
to study a promoter, it's possible to put a
reporter gene immediately downstream of a
known or artificial promoter and put that
construct back into the animal to see where it
the reporter becomes expressed breaking up the
regulatory DNA of a gene allows the function of
the individual parts to be examined enhancers
are generally modular-- they can be taken from
one gene and placed into another to allow it to
be expressed in a given tissue
17
Methods for Studying Regulatory DNA
DNAse footprinting (aka DNAse protection
assay) identifies the sequence of DNA bound by
a transcription factor protein binding prevents
DNA from being attacked by DNAse I otherwise
DNAse I cuts random sequences so that bands of
many sizes are found on a gel everywhere EXCEPT
where the transcription factor protects the DNA
18
DNA footprinting
19
Methods for Studying Regulatory DNA
gel mobility shift assay another technique for
showing that a protein binds to a given DNA
sequence short DNA sequence protein is mixed
so that they can bind if correct DNA is
separated by electrophoresis DNA bound to
protein is larger and slower than 'naked' DNA
D1 D2 D1prot D2prot
20
Methods for Studying Regulatory DNA
enhancer trap trick for identifying regulatory
DNA that turns genes on in a particular
pattern, either in a tissue or at a certain
time insert a reporter gene that has a basal
promoter but no enhancers into random genomic
DNA of an organism IF it inserts near an
enhancer, it can be expressed (therefore seen) in
the same pattern as that enhancer ie. if it
is a pancreatic enhancer, the reporter will only
be expressed in the pancreas if it is a
hematopoetic stem cell enhancer, it will only be
found in those very useful for finding out what
DNA controls localized gene expression
21
Methods for Studying Regulatory DNA
ectopic expression technique for making a
protein at the incorrect place in order to
study it's function make a transgenic animal
that expresses a transcription factor at a known
time or place (often identified using an
enhancer trap) often uses a transcription
factor in yeast, called Gal4 now, using that
transgenic line, put a gene that you're
interested in (such as a reporter OR a
functioning protein) under the control of the
Gal4 promoter-- expresses your protein of
interested where you want it
22
Methods for Studying Regulatory DNA
conditional mutation a gene is non-functional
only under some conditions most common--
temperature sensitive mutations (active at normal
temp, inactive at higher or lower
temperature) because many interesting genes are
reused during development, it is often
important to be able to have a gene functional
early (so that the animal lives) and inactive
late (so you can see the phenotype of the
mutation) 'floxed mice' genetic technique to
generate controlled conditional mutants 1)
requires an enhancer that directs protein
expression where you want it put a particular
viral DNA recombinase (Cre) with that
enhancer 2) make a knockin mutation with
recombination sites for Cre recombinase (loxP
DNA sequences) surrounding the gene (or
individual exon) of a gene you wish to
delete DNA mutation only occurs where Cre is
expressed- most cells are normal
23
Methods for Studying Regulatory DNA
extremely powerful technique-- allows you to make
mutations occur where and when you want them,
while otherwise functioning normally it is
similar to ectopic expression, except that you
can delete a gene function (and then look for
a phenotype) rather than express a new gene RNAi
may someday replace floxed mice-- so far, it's
not as reliable and the tissue specificity is
harder to control
24
RNA Processing
once a gene is transcribed, it still isn't a
functional protein mRNA needs to be processed
and modified before it can be used 1) introns
need to be removed 2) mature mRNA needs to be
moved to the cytoplasm 3) mRNA needs to be
translated into protein 4) proteins may need
to be processed in order to become active mRNA
refers to mature messenger RNA that reaches the
cytoplasm nRNA (nuclear RNA) refers to immature
RNA that is not processed different cell types
can process RNA molecules differently even
though they are transcribing the same gene
25
Anatomy of a mRNA
5' cap 7-methylguanosine modified nucleotide
that protects the RNA leader sequence
untranslated mRNA which is needed for ribosome
binding and recognition exon protein coding
region of a newly made mRNA kept in the mature
mRNA transcript intron 'intervening' DNA
sequence not coding for protein-- not much
known function alternative exon sequence which
is sometimes an exon and sometimes an intron--
spliced out
26
Anatomy of a mRNA
translation initiation 'AUG' codon (methionine)
required for a ribosome to start protein
synthesis stop codon one of 3 'nonsense' codons
that cause the ribosome to fall off 3'
untranslated region mRNA sequence after the stop
codon important for mRNA regulation and
stability polyA site sequence directing the
addition of a polyA tail usually includes a
transcription termination site downstream
enhancers/silencers are usually in introns rather
than exons
start codon
5' UTR
3' UTR
stop codon
exons
introns
5'
3'
5'
3'
mature mRNA transcript
AAAAAA
7 methylguanosine cap
27
Differential RNA Processing
starting with a pre-mRNA
alternative splicing could yield
or
or
or
or
spliceosome ribonucleoprotein complex composed
of small nuclear RNA (snRNA) that removes
introns via a lariat complex splicing factors
proteins which recognize the boundaries between
introns and exons-- if they are different
between cells, splicing will differ
28
Differential RNA Processing
alternative splicing can generate many different
mRNA transcripts from a single gene ie.
including or excluding a GPI tail (phospholipid
binding sequence) changes whether a protein is
soluble or bound to the cell surface a-tropomyosi
n is spliced differently in different types of
cells
Dscam adhesion molecule in flies is the champ--
est. 38,000 forms!
29
Differential mRNA Translation
even after splicing and secretion, mRNAs have to
be translated to make proteins longer a mRNA
is around, the longer it can make protein
therefore the more of that protein gets
produced 3' untranslated regions of mRNA seem to
regulate mRNA half-life some mRNAs persist
longer than others different cells or even
differently treated cells can express proteins
that extend or shorten the half-life of a mRNA
30
Differential mRNA Translation
oocytes often store many mRNAs prior to
fertilization 'activated' during
fertilization include bicoid and nanos that form
the gradients in syncitial specification many
organisms don't transcribe genes during
cleavage-- rely on mRNA found in oocytes--
they're too busy dividing/replicating to
transcribe significantly regulated by the 5' and
3' untranslated regions of the mRNA maskin
protein in Xenopus that blocks mRNA translation
by binding to the 5' and 3' UTRs Cortex and
Grauzone in flies are required to add a polyA
tail-- mRNAs are poorly translated without
that tail other blockages are specific
proteins-- nanos blocked by Smaug
31
Differential mRNA Translation
microRNAs also function as inhibitors of
translation lin-14 and lin-4 are two mutations
controlling larval gene expression in C
elegans lin-14 makes a protein important for
early larval organs mRNA for the gene is seen
long after the protein has disappeared lin-4 is
only 25 or 65 bp long that has no open reading
frame (ie. no protein coding sequence) but the
RNA itself is nearly identical to the 3' UTR
of lin-14 binding of lin-4 RNA to the lin-14
mRNA prevents its translation 200 of these
small inhibitory RNAs are believed to exist
naturally processed by the enzyme Dicer,
similar to what is seen in RNAi
32
Differential mRNA Translation
other transcripts are localized within the
cytoplasm bicoid and nanos in Drosophila eggs
are a common example Vg1 in Xenopus eggs is
another-- localized to the vegetal (yolk)
half in neurons, certain mRNAs are localized and
translated in dendrites calcium/calmodulin
(CaM) kinase II is there-- regulated by it's 3'
UTR putting the 3' UTR onto b galactosidase
localizes its message to dendrites
SR stratum radiale - very few cell bodies, many
dendrites (hippocampus)