molecular evolution - PowerPoint PPT Presentation
1 / 33
Title:
molecular evolution
Description:
Hoatzin (Opisthocomus hoatzin) eats leaves. only avian foregut fermenter. stomach lysozyme ... characteristic hoatzin mammalian egg-white. low pH optimum ... – PowerPoint PPT presentation
Number of Views:53
Avg rating:3.0/5.0
Title: molecular evolution
1
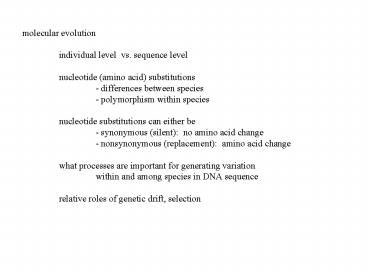
molecular evolution individual level vs.
sequence level nucleotide (amino acid)
substitutions - differences between species -
polymorphism within species nucleotide
substitutions can either be - synonymous
(silent) no amino acid change - nonsynonymous
(replacement) amino acid change what
processes are important for generating
variation within and among species in DNA
sequence relative roles of genetic drift,
selection
2
estimating substitution rates substitution
rate differences/years since divergence
mouse-human divergence 80 million years ago
protein composed of 100 amino acids - 16 differ
betw spp subst. rate 16/(100 x 2 x 80
x106) 1 X 10-9 (substitution rate of
DNA sequences more complicated because of
multiple hits due to 4 nucleotides)
3
Neutral theory of molecular evolution (Kimura
1983) advantageous mutations are rare most
sequence variation is neutral with respect to
fitness most genes evolve by genetic drift not
selection not pan-neutral, deleterious
substitutions eliminated by selection,
advantageous substitutions fixed by
selection contrast with Selectionist
theory -- natural selection is responsible
for all sequence variation
4
mutation---gtamino acid substitution had
increased to fixation every 2 years too
high to be due to natural selection
constant rate of amino acid substitution molec
ular clock -- inconsistent with change
by selection
5
Neutral theory of evolution (Kimura
1983) clock-like pattern of substitution at
amino acid or sequence level better
approximated by drift than selection in a
population of N individuals, with a mutation rate
of n, 2Nn new alleles occur each
generation the probability of fixation by drift
is 1/2N the number of new alleles that will
fix by drift is 2Nn x (1/2N) n
(substitutions/generation) over large time
scales absolute time relative frequency
of advantageous, deleterious, neutral mutations
6
nearly neutral model (Ohta 1992) genetic drift
and selection drift fixes alleles (at random)
when Ne is small directional selection fixes the
favored allele when Ne is large when does
drift overcome selection? 1/4Nes lt 1 many
mutations are effectively neutral (fixed or
eliminated by drift not selection)
7
nearly neutral model rate of neutral mutation,
n, should vary as a function of generation
time, not absolute time species with short
generation times should accumulate mutations
faster than those with long generation times
(esp. nonsynonymous subst.) but, mutations are
effectively neutral when 1 lt 4Nes
8
Chao and Carr 1993
9
nearly neutral model rate of neutral mutation,
n, should vary as a function of generation
time, not absolute time species with short
generation times should accumulate mutations
faster than those with long generation
times (mutations are effectively neutral when
1 lt 4Nes) because Ne and generation time are
negatively correlated these effects cancel out
10
Patterns of DNA sequence divergence --pseudogene
s reflect neutral rate of evolution -- silent
sites evolve faster than replacement sites
most replacement substitutions are deleterious,
and will be eliminated by selection --rate of
evolution will vary among gene regions
different gene regions may be more
functionally constrained -- hemoglobin --rate
s of evolution will vary among genes functional
constraints variation in selection
11
rates of nucleotide substitution (l) for
different regions of mammalian genes and for
pseudogenes
(modified from Li 1997)
12
substitution rates in influenza
/Replacement
13
functional constraint in hemoglobin
surface V 1.35 x 10-9 aa/site/yr
2.73 x 10-9 heme pocket V 0.17 x 10-9
0.24 x 10-9
14
Patterns of DNA sequence divergence --pseudogene
s reflect neutral rate of evolution -- silent
sites evolve faster than replacement sites
most replacement substitutions are deleterious,
and will be eliminated by selection --rate of
evolution will vary among gene regions
different gene regions may be more
functionally constrained -- hemoglobin --rate
s of evolution will vary among genes functional
constraints variation in selection
15
Rates of nucleotide substitution in mammalian
protein coding genes
codons replacement synonymous
rate rate Histones H3 135
0.00 0.00 6.38 1.19 H4 101
0.00 0.00 6.12 1.32 Ribosomal proteins
S14 150 0.02 0.02 2.16 0.42
S17 134 0.06 0.04 2.69
0.53 IGF-2 179 0.57 0.11 2.01
0.37 Albumin 590 0.92 0.07 5.16
0.48 Interferons V1 166 1.47
0.19 3.24 0.66 1 159 2.38
0.27 5.33 1.24 ( 136 3.06
0.37 5.50 1.45 (from Li 1997)
16
DNA sequence data and adaptive evolution ratio
of non-synonymous to synonymous changes
17
Comparison of the rate of Nonsynonymous
(replacement) substitutions (dN) and Synonymous
(silent) substitutions (dS) higher dN/dS
ratios can be produced by - positive selection
favoring a change in function - relaxation of
purifying selection (rapid increase in popn
size) dN/dS 0.1 0.2 no evidence of
selection dN/dS 0.2 - 1 cant
distinguish between positive selection and
relaxed selection dN/dS gt 1 positive
selection
18
evolution of leptin
dN dS
2.2
Great apes
Old World monkeys
dN dS
0.21
Rodents
Artiodactyls
Benner et al. 2002 Science 296864
19
DNA sequence data and adaptive evolution ratio
of non-synonymous to synonymous changes dN/dS
ratios within and between species -
McDonald-Kreitman test
20
neutral theory predicts a constant ratio of
replacementsilent substitutions over
time i.e., RS within species RS between
species McDonald-Kreitman test Adh locus in
Drosophila melanogaster (12) simulans (6)
yakuba (12) fixed between species polymorphic
within species Replacement 7
2 Silent 17 42 plt0.006 R
29 5 significantly different ratios
21
Fay et al 2002 Nature 4151024 45 genes --
Drosophila melanogaster and D. simulans McDonald-
Kreitman test divergent polymorphic autosoma
l 0.81 0.37 p lt 10-7 X-linked 0.22 0.13 ns ove
rall 0.63 0.29 p lt 10-6 positive
selection increase in selective constraint (
population size)
22
DNA sequence data and adaptive evolution ratio
of non-synonymous to synonymous changes dN/dS
ratios within and between species -
McDonald-Kreitman test convergence of DNA
sequence
23
lysozyme breaks down bacteria cell wall ---gt
lysis saliva, blood, tears, milk ruminants
(cows etc), leaf-eating monkeys (langur) new
form of lysozyme digest stomach
bacteria bacteria digest cellulose environment
of stomach lysozyme is more acidic
24
14 lysine 21 aspartic acid 75 aspartic
acid 87 asparagine 126 lysine
cow
other ungulates
other primates
langur
14 lysine 21 aspartic acid 75 aspartic
acid 87 asparagine 126 lysine
bird
25
Hoatzin (Opisthocomus hoatzin) eats leaves
only avian foregut fermenter stomach lysozyme
26
300 my
Kornegay et al. 1994 Mol. Biol. Evol. 11921
27
Structural adaptations in stomach lysozyme
lysozyme type characteristic
hoatzin mammalian egg-white low pH optimum
- isolelectric point
6 6.2 7.7 11.2 total arginines 5
3 6 11 aginine to lysine ratio
0.63 0.27 0.67 1.83 adaptive residues
14 E/K - 21 E/K
- 75 D
- 87 N -
126 E/K -
28
DNA sequence data and adaptive evolution ratio
of non-synonymous to synonymous changes dN/dS
ratios within and between species -
McDonald-Kreitman test convergence of DNA
sequence biased codon usage
29
Codon human Drosophila E.
coli Arginine AGA 22 10 1
AGG 23 6 1 CGA
10 8 4 CGC 22 49
39 CGG 14 9 4
CGU 9 18 49 total
arginine codons 2403 506
149 total genes 195 46
149
stop codons in mammalian mtDNA
30
leucine
31
Codon Bias differential usage of codons for
specific amino acids --variation within and
among species --bias is much stronger in highly
expressed genes decreased rate of translation
(bacteria) decreased accuracy of translation
(bacteria) decreased stability of mRNA but
individual selection is weak -- interaction
effect??
32
Correlated responses in non-selected
sequences hitchhiking and selective
sweeps positive selection increases frequency
of new replacement substitutions and closely
linked sites -- reduced polymorphism backgrou
nd selection purifying (negative) selection
eliminates deleterious mutations and reduces
polymorphism in closely linked sites
33
--genomic data facilitates studying selection at
the molecular level -- silent sites evolve
faster than replacement sites due to
purifying (negative) selection -- positive
selection indicated by dN/dS gt 1 -- rate of
evolution varies among gene regions implying
functional constraints -- rate of evolution
varies among genes implying functional
constraints or variation in selection --in
large genomes (e.g., human) most molecular
evolution may be the result of drift rather
than selection