Formation of mature eukaryotic mRNA from premRNA - PowerPoint PPT Presentation
1 / 24
Title:
Formation of mature eukaryotic mRNA from premRNA
Description:
Formation of mature eukaryotic mRNA from pre-mRNA ... forms 5'-2' bond with A in the branch-point sequence to form an RNA lariat ... – PowerPoint PPT presentation
Number of Views:103
Avg rating:3.0/5.0
Title: Formation of mature eukaryotic mRNA from premRNA
1
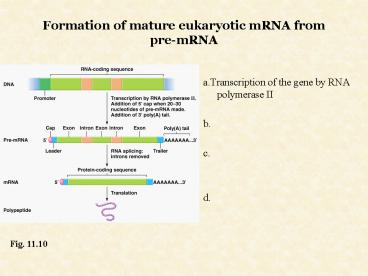
Formation of mature eukaryotic mRNA from pre-mRNA
- a.Transcription of the gene by RNA polymerase II
- b.
- c.
- d.
Fig. 11.10
2
5 Modification of Euk mRNA
- The newly made 5 end of the mRNA is modified by
5 capping - A capping enzyme adds a guanine, usually 7-methyl
guanosine (m7G), to the 5 end using a 5-to-5
linkage - Sugars of the 2 adjacent nt are also methylated
Figure 11.8 Cap structure at the 5? end of a
eukaryotic mRNA
3
3 Modification of Euk mRNA
- The 3 end of the pre-RNA has 50250 adenines
added enzymatically to form a poly(A) tail - mRNA stability
- Plays a role in transcription termination, since
RNA polymerase II does not rely directly on a
signal in the DNA
4
Fig. 11.9 Diagram of the 3? end formation of
mRNA and the addition of the poly(A) tail
- a. Transcription of mRNA continues through the
poly(A) consensus sequence (AAUAAA), the poly(A)
site and the GU-rich sequence. - b. A protein called CPSF (cleavage and
polyadenylation specificity factor) binds the
AAUAAA signal. - c. A protein called CstF (cleavage stimulation
factor) binds to the GU-rich sequence. - d. CPSF and CstF bind to each other, producing a
loop in the RNA. - e. CFI and CFII bind near the poly(A) site, and
RNA is cleaved. - f. After cleavage, the enzyme poly(A) polymerase
(PAP) - g. PABII (poly(A) binding protein II) binds the
poly(A) tail as it is produced
5
Splicing to remove introns
- Intron
- Exon
- Splicing machinery guided by conserved sequences
GU-AG rule - Pre-mRNA
- 5Exon-GUAAGU.YNCURAY.6YNCAGExon3
- Same for all nuclear mRNA genes so far
6
A Detailed look at intron removal from a
pre-mRNA molecule
7
Splicing to remove introns
- Spliceosome Nuclear complex that removes introns
and joins exons
8
Splicing to remove introns
- a. Each of the 6 principal snRNAs (named U1-U6)
is associated with 6-10 proteins to form the
snRNPs - b. Some of the proteins are specific to
particular snRNPs, and others are found in all
snRNPs - c. The U4 and U6 snRNAs occur within the same
snRNP (U4/U6 snRNP). All the other snRNPs have
only a single snRNA - d.
9
Fig. 11.11 Model for intron removal by the
spliceosome
- snRNP U1
- snRNP U2
- snRNPs U4, U5, and U6 interact and bind U1 U2
forming intron loop - snRNP U4 dissociates from the complex, forming
the active spliceosome - Intron cut at 5 G of intron/exon junction
- 5 G end of intron forms 5-2 bond with A in the
branch-point sequence to form an RNA lariat - Intron cut at 3 end G of AG
- Intron is released and degraded
- Exons 1 and 2 are ligated
- snRNPs are released
10
RNA Editing
- Posttranscriptional insertion, deletion, or
chemical alteration of bases - of nucleotides from a pre-mRNA
- Resulting mRNA has bases that dont match its DNA
coding sequence - 2. Examples of RNA editing have been found in a
number of organisms - a. In Trypanosome brucei (a protozoan causing
sleeping sickness) the cytochrome oxidase subunit
III gene (from mitochondrial DNA) does not match
its mRNA. Uracil residues have been added and
removed, and over 50 of the mature mRNA consists
of posttranscriptionally added Us. This RNA
editing is mediated by a guide RNA (gRNA) that
pairs with the mRNA, cleaving it, adding the Us
and ligating it
Fig. 11.12 Comparison of the DNA sequences of
the cytochrome oxidase subunit III gene in the
protozoans Trypanosome brucei (TB), Crithridia
fasiculata (Cf), and Leishmania tarentolae
(Lt), aligned with the conserved mRNA for Tb
11
Transcription of Other Genes
- Genes that do not encode proteins are also
transcribed, including genes for rRNA, tRNA and
snRNA
12
Ribosomal RNA and Ribosomes
- Ribosomes are the catalyst for protein synthesis
(translation) - Facilitating binding of charged tRNAs to the mRNA
so that peptide bonds can form - A cell contains thousands of ribosomes
- Comprised of small and large subunits
- Each subunit has
13
Ribosomal RNA and Ribosomes
- Prokaryotes
- 50S large subunit
- 30S small subunit
- Subunits interlock to form
- Three rRNAs
14
The E. coli Ribosome
Before translation can occur, subunits need to
assemble themselves A complete ribosome
is formed when subunits interlock when attaching
to mRNA during initiation of translation
15
30S Small subunit
3D shape allows subunits to interlock
16
Transcription of prokaryotic rRNA Genes
- DNA regions that encode rRNA are called ribosomal
DNA (rDNA) or rRNA transcription units. - E. coli is a typical prokaryote, with
- a. Each rrn contains the rRNA genes 16S-23S-5S,
in that order, with tRNA sequences in the
spacers. - b. A single pre-rRNA transcript is produced from
rrn, and cleaved by RNases to release the rRNAs.
Cleavage occurs in a complex of rRNA and
ribosomal proteins, resulting in functional
ribosomal subunits.
17
Fig. 11.15 rRNA genes and rRNA production in E.
coli
18
Ribosomal RNA and Ribosomes
- Eukaryotes
- 60S
- 40S
- Interlock to form
- Larger and more complex than prokaryotic
subunits, and vary in size and composition among
organisms - 3 to 5 diff rRNAs (5S, 5.8S, 18S, 28S in
mammals)
19
The Mammalian Ribosome
20
Transcription of eukaryotic rRNA Genes
- The three rRNA genes with homology to prokaryotic
rRNA genes are 18S-5.8S-28S, in that order. In
the chromosome these genes are tandemly repeated
1001,000 times to form rDNA repeat units. The 5S
rRNA gene copies are located elsewhere in the
genome. - RNA polymerase I transcribes the rDNA repeat
units, producing a single large pre-rRNA molecule
containing the 18S, 5.8S and 28S rRNAs, separated
by ...
21
Fig. 11.16 rRNA genes and rRNA production in
eukaryotes
- Specific cleavage steps free the rRNAs from their
transcript as part of pre-rRNA processing that
takes place in the complex of pre-rRNA, 5S rRNA
and ribosomal proteins. The result is formation
of 40S and 60S ribosomal subunits, which are then
transported to the...
22
Transcription of rRNA in euks
- A nucleolus forms around each rDNA repeat unit,
and then they fuse to make one nucleolus.
Ribosomal subunits are produced in the nucleolus
by addition of the 5S rRNA and ribosomal
proteins.
23
Eukaryotic Transcription of rRNA
Nucleolus, site for RNA pol I transcription r
RNA precursor processing ribosome assembly
24
Fig. 11.17 Self-splicing reaction for the group
I intron in Tetrahymena pre-rRNA
- The intron self-splices by folding into a
secondary structure that - Discovery of ribozymes (catalytic RNAs) has
significantly altered our view of the
biochemistry involved in the origin of life