I'Introduction to PTMs - PowerPoint PPT Presentation
1 / 24
Title:
I'Introduction to PTMs
Description:
Large-scale phosphorylation site mapping ... Immobilized metal affinity chromatography (IMAC) ... Introduce affinity tag to enrich for phosphorylated molecules ... – PowerPoint PPT presentation
Number of Views:96
Avg rating:3.0/5.0
Title: I'Introduction to PTMs
1
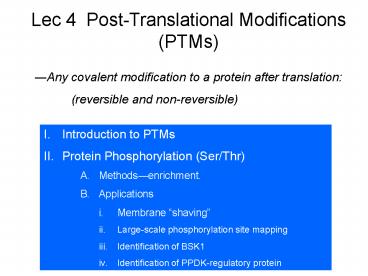
Lec 4 Post-Translational Modifications (PTMs)
?Any covalent modification to a protein after
translation (reversible and non-reversible)
- I. Introduction to PTMs
- Protein Phosphorylation (Ser/Thr)
- Methods?enrichment.
- Applications
- Membrane shaving
- Large-scale phosphorylation site mapping
- Identification of BSK1
- Identification of PPDK-regulatory protein
2
Mechanisms by which a single gene can give rise
to multiple gene products.
Today, more than 200 modifications are known to
occur. 5-10 of the genome estimated to encode
enzymes that catalyze modifications.
3
Proteomic analysis of PTMs
Mann and Jensen, Nature Biotech. 21, 255 (2003)
4
Reversible Protein Phosphorylation Regulates Most
Aspects of Cell Life
1992 Nobel Prize to Fischer and Krebs
5
Reversible Protein Phosphorylation
OGT O-GlcNAc Transferase, encoded by SPY
6
- Protein Phosphorylation
- One of the most common intracellular protein
modification - Approximately 5 of the Arabidopsis thaliana
genome encodes for protein kinases phosphatases
(H. sapiens 5 ) - Approximately 30 of all human proteins are
phosphorylated. How about plants, e.g., A.
thaliana? - Protein phosphorylation is of special importance
in the regulation of functions, e.g. metabolism
or cell signaling
In order to better understand the molecular
mechanisms where phosphorylation is involved this
modification has to be analyzed.
7
Methods for Phosphorylation Analysis
- For selective detection
- Radioactive labeling with 32P or 33P
- Western blotting with anti-phosphoamino acid
antibodies - ProQ-Diamond Stain
- For separation
- Liquid chromatography (LC)
- Thin layer chromatography (TLC)peptide mapping
- (Gel) Electrophoresis
- For exact localization
- Edman sequencing
- Site-directed mutagenesis
- Mass spectrometry
8
MS-Based Phosphorylation Analysis
- Enrichment of phosphorylated proteins/peptides
- Anti-phosphoamino acid antibodies
- IMAC
- Derivatization
- 2) localization of the phosphorylation site
- MALDI-ToF-MS
- LC-MS/MS (e.g., LTQ-Orbitrap)
9
Enrichment strategies to analyze
phosphoproteins/peptides
- Phosphospecific antibodies (example coming)
- Anti-pY quite successful
- Anti-pS and anti-pT not as successful, but may
be used - (M. Grønborg, et al. Approach for Identification
of Serine/Threonine-phosphorylated Proteins by
Enrichment with Phospho-specific Antibodies.
Mol. Cell. Proteomics 2002, 1517527. - Immobilized metal affinity chromatography (IMAC)
- Negatively charged phosphate groups bind to
postively charged metal ions (e.g., Fe3, Ga3)
immobilized to a chromatographic support - Limitation non-specific binding to acidic side
chains (D, E) - Derivatize all peptides by methyl esterification
to reduce non-specific binding by carboxylate
groups. - Ficarro et al., Nature Biotech. (2002), 20, 301.
10
Enrichment strategies to analyze
phosphoproteins/peptides
Chemical derivatization Introduce affinity tag
to enrich for phosphorylated molecules e.g.,
biotin binding to immobilized avidin/streptavidin
- Oda et al., Nature Biotech. 2001, 19, 379 for
analysis of pS and pT
11
Enrichment of phosphoproteins using
anti-phosphoThr and anti-phosphoSer Abs.
12
Phosphorylation Analysis by MS The Problems
1) Ionization suppression
Phosphopeptides do not fly well and their
ionization can be suppressed by nearby (more
abundant) ions. Also low stoichiometry and loss
due to phosphatase action.
13
- Applications
- Neat techniquemembrane shaving
- Large-scale phosphorylation mapping
(LTQ-Orbitrap) - Identification of BSK1
- Identification of PPDK-RP
14
Protein Profiling by Shotgun Proteomics
- Examples could include
- chloroplast proteins
- vacuolar proteins
- cell wall proteins
- seed proteins
- membrane phosphoproteins
Resing and Ahn (2005) Proteomics strategies for
protein identification. FEBS Lett 579 885-889.
15
MEMBRANE SHAVING
Mol. Cell Proteomics 2 (2003) 1234-1243.
- Use of Brij-58 to invert PM vesicles as
demonstrated by ATP-dependent proton pumping. - Bacterial virulence factors increase in vivo
phosphorylation of PM proteins (upward arrows
TLC peptide mapping).
16
In Vivo Phosphorylated Membrane Proteins
2-dimensional LC separation of proteins.
Enrichment of phosphopeptides by IMAC step is
apparent.
Multiply phosphorylated peptides tend to elute at
high salt, as expected.
17
Novel Phosphorylation Sites on H-ATPases
MS/MS spectra for two phosphopeptides
18
Novel Phosphorylation Sites on H-ATPases
Known site, 14-3-3 protein binding site
(activates pump).
New Sites, Unknown function
19
Non-phosphorylated contaminants from IMAC
represent abundant rather than acidic peptides
(Figure 7)
20
AHA2
large-scale strategy for identification (by mass
spectrometry) of phosphorylation sites in
integral PM proteins membrane-shaving of tryptic
fragments. Identified more than 300
phosphorylation sites!
21
Phosphorylation sites of Receptor-Like Kinases
22
- CONCLUSIONS
- Phosphorylation sites tend to cluster outside
known functional domain. - Identification of a phosphopeptide establishes
the corresponding loop as cytoplasmic (membrane
topology). - Conservation of phosphorylation sites in paralogs
(isoforms) and orthologs (species) gives insights
to isoform-specific regulation. - Database resource for the Plant Biology
community. (http//plantsp.sdsc.edu) - Regulation by phosphorylation may be complex
(e.g., RLKs).
23
Common themes in plant and animal receptor
protein kinases
(Shiu and Bleecker. Science's STKE 113 (2001) 22.)
24
Representative members of the receptor-like
kinase (RLK) family.
Clavata1 is 10 BRI1 is 11. The LRR-RLKs account
for nearly half of the total RLKs (235/622). The
next largest class is the sugar-binding motif or
lectins. Function of only a few are known.