Two steps of hierarchical clustering - PowerPoint PPT Presentation
1 / 25
Title:
Two steps of hierarchical clustering
Description:
Advantages and Disadvantages of Hierarchical clustering. Advantages: 1) Straightforward ... What are the disadvantages of k-means clustering? 23 ... – PowerPoint PPT presentation
Number of Views:101
Avg rating:3.0/5.0
Title: Two steps of hierarchical clustering
1
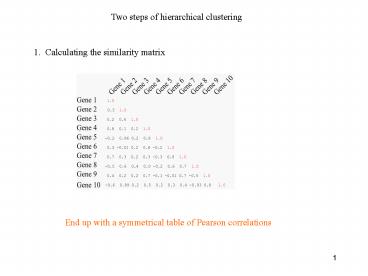
Two steps of hierarchical clustering
1. Calculating the similarity matrix
End up with a symmetrical table of Pearson
correlations
2
centroid (average vector)
4. Centroid linkage clustering
3
Visualization Data are often converted to a
colorimetric scale
Each box a transcript measurement Each row of
boxes transcript measurements for a given
gene Each column of boxes transcript
measurements from a single array Red higher
transcript abundance in one sample Green
higher transcript abundance in the other
sample
4
Software for clustering and visualization
Cluster (Mike Eisen) http//rana.lbl.gov (for PC
only) Cluster (de Hoon) http//bonsai.ims.u-tok
yo.ac.jp/mdehoon/software/cluster/ Java
Treeview (Alok Saldana) http//jtreeview.sourcef
orge.net/
5
Unweighted Pearson correlation
6
Sometimes, want to use the weighted pearson
correlation
N
1 (Xi) (Yi)
S
S x,y
N
N
i 1
1
N
2
1
S
2
S
Xi
Yi
N
N
i 1
i 1
Array 1
Array 2
Array 3
Array 4
Array 5
Gene X X1 X2 X3 X4 X5
Gene Y Y1 Y2 Y3 Y4 Y5
For example if these arrays are identical, the
data are over-represented 3X
7
Sometimes, want to use the weighted pearson
correlation
N
1 (Xi) (Yi)
S
S x,y
wi
wi
S
N
i 1
1
N
2
1
S
2
S
Xi
Yi
N
N
i 1
Where wi 1 Li
Array 1
Array 2
Array 3
Array 4
Array 5
Gene X X1 X2 X3 X4 X5
k array corr. cutoff d Pearson distance ( 1
- P. corr) n exponent (usually 1)
Gene Y Y1 Y2 Y3 Y4 Y5
For example if these arrays are identical, the
data are over-represented 3X -- can weight
experiments i 3,4,5 by w 0.33
8
Unweighted Pearson correlation
Weighted Pearson correlation
9
Unweighted Pearson correlation
Weighted Pearson correlation
10
Can also cluster array experiments based on
global similarity in expression
Alizadeh et al. 2000
11
Hierarchical trees of gene expression data are
analogous to phylogenetic trees
A
D
B
Distance between genes is proportionate to the
total branchlength between genes (not the
distance on the y-axis)
E
F
C
Orientation of the nodes is irrelevant
. although some clustering programs try
to organize nodes in some way.
12
Hierarchical trees of gene expression data are
analogous to phylogenetic trees
A
D
B
Distance between genes is proportionate to the
total branchlength between genes (not the
distance on the y-axis)
E
F
C
Orientation of the nodes is irrelevant
. although some clustering programs try
to organize nodes in some way.
D
B
A
E
F
C
13
Advantages and Disadvantages of Hierarchical
clustering
Advantages 1) Straightforward 2) Captures
biological information relatively
well Disadvantages 1) Doesnt give
discrete clusters need to define clusters with
cutoffs 2) Hierarchical arrangement does not
always represent data appropriately --
sometimes a hierarchy is not appropriate genes
can belong only to one cluster.
14
(No Transcript)
15
(No Transcript)
16
(No Transcript)
17
(No Transcript)
18
(No Transcript)
19
Advantages and Disadvantages of Hierarchical
clustering
Advantages 1) Straightforward 2) Captures
biological information relatively
well Disadvantages 1) Doesnt give
discrete clusters need to define clusters with
cutoffs 2) Hierarchical arrangement does not
always represent data appropriately --
sometimes a hierarchy is not appropriate genes
can belong only to one cluster. 3) Get
different clustering for different experiment
sets THERE IS NO ONE PERFECT CLUSTERING METHOD
20
k-means clustering
Partitioning (or top-down) clustering method --
Randomly split the data into k groups of equal
number of genes -- Calculate the centroid of
each group -- Reassign genes to the centroid to
which it is most similar -- Calculate a new
centroid for each group, reassign genes, etc
iterate until stable
21
k-means clustering
Partitioning (or top-down) clustering method --
Randomly split the data into k groups of equal
number of genes -- Calculate the centroid of
each group -- Reassign genes to the centroid to
which it is most similar -- Calculate a new
centroid for each group, reassign genes, etc
iterate until stable
Centroids
22
k-means clustering
Partitioning (or top-down) clustering method --
Randomly split the data into k groups of equal
number of genes -- Calculate the centroid of
each group -- Reassign genes to the centroid to
which it is most similar -- Calculate a new
centroid for each group, reassign genes, etc
iterate until stable
What are the disadvantages of k-means clustering?
23
k-means clustering
Partitioning (or top-down) clustering method --
Randomly split the data into k groups of equal
number of genes -- Calculate the centroid of
each group -- Reassign genes to the centroid to
which it is most similar -- Calculate a new
centroid for each group, reassign genes, etc
iterate until stable
What are the disadvantages of k-means clustering?
- Need to know how many clusters to ask for
- (can define this empirically)
- Genes are not organized within each cluster
- (can hierarchically cluster genes afterwards or
use SOM analysis) - - Random process makes this an indeterminate
method
24
Brief overview of other organizational methods
Principal Component Analysis (PCA) Singular
Value Decomposition (SVD) -- reduce data to a
series of representative expression patterns
(eigen genes) the together summarize the
data - principal component summarizes the
majority of the data - secondary components
summarize minor components of data - real genes
some sum of components
Bayesian approaches Probabalistic modeling of
gene expression data Support Vector Machines
(SVM) series of lines that partition the data
into subgroups
25
What kinds of information can we extract from
whole-genome expression data?
- Hypothetical functions for uncharacterized genes
- -- genes encoding subunits of multi-subunit
protein complexes - are often highly coregulated
- example ribosomal protein genes, proteasome
genes in yeast - -- genes involved in the same cellular processes
are often coregulated
- New roles for characterized genes
- Better understanding of the experimental
conditions - -- based on expression patterns of characterized
genes - Implications of gene regulation
- -- WT vs. mutants can identify transcription
factor targets - -- promoter analysis of coregulated genes
upstream elements - -- gene coregulation with known pathway targets
can implicate - pathway activity
- Understanding developmental pathways
- Defining experimental samples based on expression
profiles - example comparing tumor samples from patients