Phenotypic variation in human height - PowerPoint PPT Presentation
1 / 25
Title:
Phenotypic variation in human height
Description:
Several species that are pollinated differently. Bee pollinated (Shallow, yellow, ... Parental genotype was essentially homozygous for floral morphology. ... – PowerPoint PPT presentation
Number of Views:84
Avg rating:3.0/5.0
Title: Phenotypic variation in human height
1
- Phenotypic variation in human height
2
Phenotypic variation in Human Swimming speed
3
(No Transcript)
4
(No Transcript)
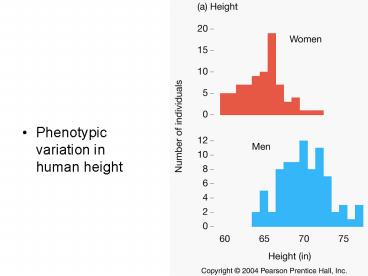
5
Note the near perfect bell-shaped curve. So, by
the central limit theorem from probability, the
sum of 6 binomially distributed random
variables is already normally distributed.
6
Note the recovery of parental phenotypes in the
F5 generation even though all members of F1 were
heterozygotes.
7
Effects of environment on phenotype. So, in the
context of our previous discussion, some of this
difference is a result of Ve
8
Monkey Flowers
- Several species that are pollinated differently
- Bee pollinated (Shallow, yellow, landing strip)
- Hummingbird pollinated (Tubular, red, nectar)
- Reproductively isolated in the field.
- Easily hybridized in the lab.
9
(No Transcript)
10
Monkey Flowers
- Note the ancestral condition is a bee pollinated
flower. - Are the alleles that cause these differences
subtle in their effects, or are they obvious in
their effects?
11
QTL Mapping in Monkey Flowers
- Selected 66 possible marker loci.
- Parental genotype was essentially homozygous for
floral morphology. - Crossed the 2 species in the lab to produce F1
hybrids. - Crossed the F2s to produce 465 individuals.
12
- The logic behind QTL mapping
13
Notice the bars on the right of each graph these
QTLs explain a considerable amount of the
variation in the phenoptypic variance.
14
Likelihood ratios
- General statistical procedure.
- In our case
- Imagine 2 scenarios
- QTL and marker locus that are on different
chromosomes, or recombine in an unlinked fashion. - QTL and marker locus that are linked, and
crossover only 10 of the time.
15
Likelihood ratios
- With linkage set at 0.1, the expected gametes
from an F1 individual that is MP/mp would be - MP 45
- Mp 5
- mP 5
- mp 45
- So, the probability of getting an MP/MP offspring
would be (.45)(.45) .2025
16
Likelihood ratios
- With linkage set at 0.5 (free recombination), the
expected gametes from an F1 individual that is
MP/mp would be - MP 25
- Mp 25
- mP 25
- mp 25
- And the proportion of MP/MP offspring would be
(.25)(.25) .0625
17
Likelihood ratios
- To get the likelihood ratio, divide the liklehood
of the linkage scenario by the likelihood of the
free recombination scenario. - 0.2025/0.0625 3.24
- So, the double homozygote is 3 times more likely
under the linkage model than under the free
recombination model.
18
Likelihood ratios
- Next, we need to do this for multiple
individuals, or across an entire population. - Here, we take the log of the likelihood ratio to
produce a LOD score (log of the odds). - Imagine we have 10 individuals with the following
genotypes (and their likelihood ratios) - 3 MMPP 3 x .511
- 4 MmPp 4 x .215
- 1 Mmpp 1 x -.444
- 2 mmpp 2 x .511
- The total sum is then 2.97
- Generally, If this value is 3.0 or larger, we say
the trait locus and the marker are linked.
19
(No Transcript)
20
(No Transcript)
21
(No Transcript)
22
(No Transcript)
23
(No Transcript)
24
(No Transcript)
25
Selection Gradient and Selection Differential
- Selection Differential Difference between mean
for a particular train among the survivors and
the mean for the entire population. - Selection Gradient Selection differential
divided by the variance of the trait. We can
show easily that this is really the slope of the
regression of fitness on the trait.