Hierarchical%20Bayesian%20Model%20Specification - PowerPoint PPT Presentation
Title:
Hierarchical%20Bayesian%20Model%20Specification
Description:
Model is specified by the Directed Acyclic Network (DAG) and the conditional ... 5685 Yeast Genes Across Two Experiments (Cell Cycle and Sporulation) ... – PowerPoint PPT presentation
Number of Views:77
Avg rating:3.0/5.0
Title: Hierarchical%20Bayesian%20Model%20Specification
1
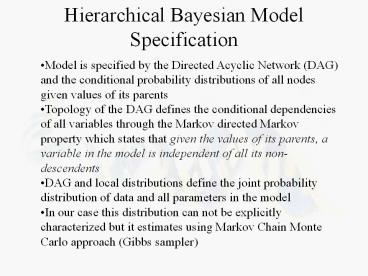
Hierarchical Bayesian Model Specification
- Model is specified by the Directed Acyclic
Network (DAG) and the conditional probability
distributions of all nodes given values of its
parents - Topology of the DAG defines the conditional
dependencies of all variables through the Markov
directed Markov property which states that given
the values of its parents, a variable in the
model is independent of all its non-descendents - DAG and local distributions define the joint
probability distribution of data and all
parameters in the model - In our case this distribution can not be
explicitly characterized but it estimates using
Markov Chain Monte Carlo approach (Gibbs sampler)
2
Uses and Miss-Uses of Clustering
- Define a statistical model that facilitates
clustering of genes based on similarities of
their expression profiles - Define the method-selection criteria that allows
for estimating the "correct" number of clusters - Show that inappropriate "pre-filtering" can fool
the statistical model in the same way it fools
the casual observer - Show appropriate ways to use cluster analysis and
illustrate the importance of using the "best
available treatment"
3
Clustering of gene expression profiles
4
Patterns of Expression - Finite Mixture Model
Patterni ? ?i(?1i, ?2i,, ?11i) Dataik iid
N(?i, ?), k1,,ni ninumber of genes generated
by the Patterni ?ini/n
5
Patterns of Expression - Finite Mixture Model
Any gene profile x (x1,x2,,x11)
Finite Mixture Model
?
All data x1, x2,, xn
6
One-dimensional mixture
Pattern 1
N(?11, ?)
N(?12, ?)
Pattern 2
7
MCLUST
gt library(mclust) gt SimDatalt-matrix(rnorm(500015)
,ncol15) gt ColLabelslt-c(paste("Tumor_",18,sep""
),paste("Control_",17,sep"")) gt
heatmap(SimData,labColColLabels) gt
.MclusthcModelNameslt-c("E","EEI") gt
.MclustemModelNameslt-c("EEI") gt
BIC.emclustlt-EMclust(SimData,110) gt
BIC.emclust BIC EEI 1 -213490.3 2
-213624.9 3 -213753.0 4 -213880.7 5
-213993.7 6 -214121.0 7 -214243.4 8
-214351.6 9 -214481.4 10 -214588.7 gt
plot(BIC.emclust) EEI "1" gt
8
Determining the number of patterns
9
MCLUST
gt p.valuelt-apply(SimData,1,function(x)
t.test(x18,x915,var.equalT)p.value) gt gt
SigDatalt-SimDatap.valuelt0.05, gt
dim(SigData) 1 242 15 gt heatmap(SigData,labCol
ColLabels) gt gt BIC.emclustlt-EMclust(SigData,110)
gt BIC.emclust BIC EEI 1
-10599.485 2 -9647.645 3 -9685.897 4
-9729.239 5 -9796.119 6 -9849.109 7
-9912.601 8 -9973.645 9 -10037.436 10
-10077.862 gt plot(BIC.emclust) EEI "1" gt
10
Determining the number of patterns
11
Summary
- The "weak filter" based on selecting
"sub-significant" differentially expressed genes
created artificial clusters - When the whole dataset was used, the Bayesian
information criteria did the right thing by
estimating the correct number of clusters to be
equal to one - Take home message When "filtering" before
clustering make sure that appropriate statistical
significance levels have been used
12
Using clustering to find "patterns" among
differentially expressed genes
- Cluster analysis is preceded by a rigorous
statistical analysis - For example-identify genes that were
"differentially" expressed on at least one
experimental comparison. Among all these genes
some will have similar behavior across all
experimental conditions - Clustering is a way of organizing behavior of
differentially expressed genes across different
experimental conditions
13
Using clustering to find "patterns" among
differentially expressed genes
14
Using clustering to find "patterns" among all
genes
- No filtering is performed
- You can perform the "quality filtering"
- Trying to identify statistically significant
patterns - Using the best available method becomes extremely
important
15
Does It matter which clustering procedure we use?
- 5685 Yeast Genes Across Two Experiments (Cell
Cycle and Sporulation) - NO VARIABILITY BASED FILTER
- 135 Genes with closest co-expression partners
Simple Commonly Used Method (Euclidian Distance
Based Hierarchical Clustering)
"Complicated" Method (Context-specific Infinite
Mixtures)
16
"Objective" Performance Assessments Using KEGG as
the Gold Standard
- Due to a large imbalance between the total number
of negative and positive pairsThere are 17
times more negative pairs than positive pairs - a
small FPR can still produce more false positive
than true positives
17
Summary
- Using clustering alone, one can identify
"significant" patterns of expression when using
appropriate methodology - For example, Yeast data clustered in this example
did not have any replicates so the traditional
analysis to identify differentially expressed
genes before clustering is not feasable - Statistical significance of resulting clusters
needs to be carefully examined
18
Infinite Bayesian Mixtures
?
M(?1,, ?K) ?(?1,, ?K) ?(?1,,
?K) C(c1,,cN) ci?1,, K
?
?
r
?
w
C
?
M
X
19
Conditional posterior distributions and Gibbs
Sampler
20
Gibbs Sampler Result
Sequence (ck,1,.,ck,n), k1,,kmax such that
- Posterior distribution summarized through
posterior pairwise probabilities of
co-expression p(cicjX)
21
Properties
- Pooling information from the whole dataset by
estimating both patterns and assignments
similar to K-means (K-means is actually
equivalent to a special case of the mixture
models with known number of clusters) - Does not require specification of the right
number of clusters (unlike K-means) - Gives direct estimates of statistical
significance (unlike anything else on the market) - Instead of lamenting which distance measure to
use focus on the appropriate statistical model
which is a well-defined problem - Works for any type of data
22
Finding important functional groups for
up-regulated genes
Using the "Ease" annotation tool
http//david.niaid.nih.gov/david/ We obtained
following significant gene ontologies Up_DexANDNE2
ANDirr_381_GO.htm Homework 1) Download and
install Ease 2) Select top 20 most-signficianly
up-regulated genes in our W-C dataset and
identify significantly over-represented
categories (using the three-way ANOVA
analysis) 3) Repeat the analysis with 30, 40, 50
and 100 up-regulated and down-regulated gene 4)
Prepare questions for the next class regarding
problems you run into