Objective - PowerPoint PPT Presentation
1 / 1
Title:
Objective
Description:
PEs form Coterie based upon some condition, dictated during execution of algorithm ... A 5 x 5 Coterie Network. Results. Constant time algorithm for exact match. ... – PowerPoint PPT presentation
Number of Views:31
Avg rating:3.0/5.0
Title: Objective
1
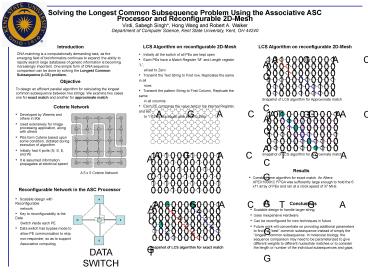
Solving the Longest Common Subsequence Problem
Using the Associative ASC Processor and
Reconfigurable 2D-Mesh Virdi, Sabegh Singh, Hong
Wang and Robert A. Walker Department of Computer
Science, Kent State University, Kent, OH 44240
LCS Algorithm on reconfigurable 2D-Mesh
LCS Algorithm on reconfigurable 2D-Mesh
Introduction DNA matching is a computationally
demanding task, as the emerging field of
bioinformatics continues to expand the ability
to rapidly search large databases of genetic
information is becoming increasingly important.
One simple form of DNA sequence comparison can be
done by solving the Longest Common Subsequence
(LCS) problem.
- Initially all the switch of all PEs are kept
open - Each PEs have a Match Register M and Length
register L, - all set to Zero
- Transmit the Text String to First row,
Replicates the same in all - rows
- Transmit the pattern String to First Column,
Replicate the same - in all columns
- Each PE compares the value held in his
internal Register, and set - to 1 if they are equal, else set it to Zero
Objective To design an efficient parallel
algorithm for calculating the longest common
subsequence between two strings. We examine two
cases one for exact match and another for
approximate match
Snapshot of LCS algorithm for Approximate match
Coterie Network
- Developed by Weems and others in 90s
- Used extensively for Image processing
application, along with others - PEs form Coterie based upon some condition,
dictated during execution of algorithm - Initially had 4 ports (N, S, E, and W)
- It is assumed information propagates at
electrical speed
- Results
- Constant time algorithm for exact match. An
Altera APEX1000KC FPGA was sufficiently large
enough to hold the 6 x11 array of PEs and ran at
a clock speed of 37 MHz.
A 5 x 5 Coterie Network
Reconfigurable Network in the ASC Processor
- Scalable design with Reconfigurable
- network
- Key to reconfigurability is the Data
- Switch inside each PE
- Data switch has bypass mode to
- allow PE communication to skip
- non-responder, so as to support
- Associative computing
- Conclusions
- Scalable design to handle larger string
- Uses Inexpensive Hardware
- Can be reconfigured for new techniques in future
- Future work will concentrate on providing
additional parameters to find the best common
subsequence instead of simply the longest
common subsequence. In molecular biology, the
sequence comparison may need to be parameterized
to give different weights to different nucleotide
matches or to consider the length or number of
the individual subsequences and gaps.
Snapshot of LCS algorithm for exact match