ChIP DNA Sample Preparation - PowerPoint PPT Presentation
1 / 5
Title:
ChIP DNA Sample Preparation
Description:
that anneal to the ends of the adapters. Amplify using the following PCR protocol: ... Primer Annealing. 3'-ATCAGAACCTAGCTG-5'P. P5'-AGTCTTGGATCGACA-3' 3' ... – PowerPoint PPT presentation
Number of Views:46
Avg rating:3.0/5.0
Title: ChIP DNA Sample Preparation
1
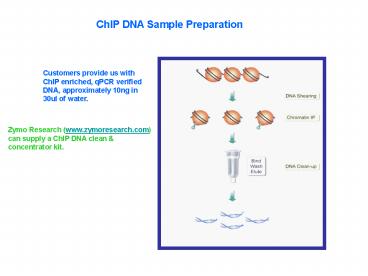
ChIP DNA Sample Preparation
Customers provide us with ChIP enriched, qPCR
verified DNA, approximately 10ng in 30ul of water.
Zymo Research (www.zymoresearch.com) can supply a
ChIP DNA clean concentrator kit.
2
ChIP DNA Sample Preparation
2. Perform End Repair
5-AGTCTTGGATCGACTCT-3
3-ACCTAGCTG-5
Convert the overhangs from into phosphorylated
blunt ends, using T4 polymerase E. coli
DNA Polymerase I large fragment (Klenow
polymerase), T4 polynucleotide kinase (PNK).
P5-AGTCTTGGATCGAC-3 3-TCAGAACCTAGCTG-5P
The 3 to 5 exonuclease activity of these
enzymes removes 3 overhangs the polymerase
activity fills in the 5 Overhangs.
3. Add A Bases to the 3 End of the DNA
Fragments
P5-AGTCTTGGATCGAC-3 3-TCAGAACCTAGCTG-5P
An A base is added to the 3 end of the blunt
phosphorylated DNA fragments, using the
polymerase activity of Klenow fragment (3 to 5
exo minus). This prepares the DNA fragments for
ligation to the adapters, which have a single T
base overhang at their 3 end
P5-AGTCTTGGATCGACA-3 3-ATCAGAACCTAGCTG-5P
3
ChIP DNA Sample Preparation
4. Ligate Adapters to the DNA Fragments
P5-AGTCTTGGATCGACA-3 3-ATCAGAACCTAGCTG-5P
Adapters are ligated to the ends of the DNA
fragments, preparing them to be hybridized to the
flow cell
P5-AGTCTTGGATCGACA-3 3-ATCAGAACCTAGCTG-5P
5. Purify Ligation Products
The products from the ligation are purified on
a 2 agarose gel, to remove all unligated
adapters, Remove any adapters that may have
ligated to one Another, select a size range of
templates to go on The cluster generation
platform
Excise a band in the gel from the 200 /- 25bp
range. Also excise a gel slice from an empty well
in the same size range, to be used as a negative
control.
4
ChIP DNA Sample Preparation
6. Enrich the Adapter-Modified DNA Fragments by
PCR
PCR is used to selectively enrich those
DNA fragments that have adapter molecules on both
ends, to amplify the amount of DNA in
the library. The PCR is performed with two
primers that anneal to the ends of the adapters
P5-AGTCTTGGATCGACA-3 3-ATCAGAACCTAGCTG-5P
Denaturation
P5-AGTCTTGGATCGACA-3
Amplify using the following PCR protocol 30
seconds at 980C 18 cycles of 10 seconds at
980C 30 seconds at 650C 30 seconds at
720C 5 minutes at 720C Hold at 40C
Primer Annealing
3-ATCAGAACCTAGCTG-5P
Extension
P5-AGTCTTGGATCGACA-3
3-NNNNNTCAGAACCTAGCTGTNN-5
As a negative control, perform PCR amplification
on the sample extracted from the empty well.
5-NNTAGTCTTGGATCGACNNNNN-3
3-ATCAGAACCTAGCTG-5P
5
ChIP DNA Sample Preparation
7. Validate the Library
The amount of starting material is very low
(10ng), and after 18 cycles of PCR the yield
could still be too low to see on a regular gel,
even though it is enough for cluster generation.
The more sensitive method of quality control
analysis is to run the library on an Agilent 2100
Bioanalyzer, to check the size, purity and
concentration. Can not use an OD260/280 ratio for
concentration measurements, since this will not
distinguish dsDNA from primers, and therefore
cannot be used to validate the library.
High MW Marker
Fluorescent Units (FU)
Sample
Low MW Marker
Fragment Size (bp)
Ladder
Sample