Protein Synthesis Translation mRNA processing is not shown - PowerPoint PPT Presentation
1 / 48
Title:
Protein Synthesis Translation mRNA processing is not shown
Description:
Protein Synthesis (Translation) (mRNA processing is not shown) ... tetra nucleotide ... X-ray Diffraction - Others... November 26 and 28, 2003. MBB 222 ... – PowerPoint PPT presentation
Number of Views:335
Avg rating:3.0/5.0
Title: Protein Synthesis Translation mRNA processing is not shown
1
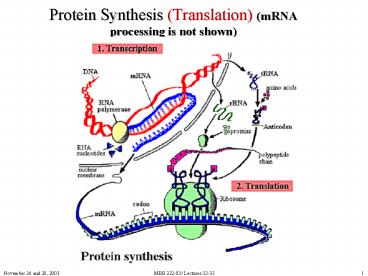
Protein Synthesis (Translation) (mRNA processing
is not shown)
2
(No Transcript)
3
Most biological activities are carried out by
proteins, and their synthesis is at the heart of
cellular function The design of protein synthesis
apparatus is similar in all organisms
RNA plays 3 Distinct Important Roles tRNA
the key or adaptor to the genetic code. rRNA
Provides scaffold catalysis. mRNA The
intermediary between gene and protein
4
Fig. 5.20
5
The Genetic Code tRNA Structure Function
Translation How is the protein coding
information in mRNA (in the language of
nucleotides) translated into a peptide
(language of amino acids)? Early
Observations - mRNA binds to ribosome - protein
is produced
Original Hypothesis Amino acids bind directly to
the nucleotides of the mRNA - by some
mutual recognition arrangement? Wrong! no
evidence, difficult to envisage from model
building
6
Alternate Hypothesis An adaptor molecule
exists
aa
5
3
tRNA
Anti-codon loop
Codon has 3 nucleotides
5
3
mRNA
The adaptor is transfer RNA (tRNA) each is
70-80 bases long. - each type of tRNA carries a
specific amino acid binds only to mRNA sites
corresponding to that amino acid (aa) - enzymes
called tRNA Synthetases recognize ONE amino acid
and its corresponding tRNAs (How? The enzymes
dont necessarily look at the sequence of the
anti codon look for more sophisticated 3-D
configurations of the tRNA)
7
Figure 27.6
In many cases, added post- transcriptionally
- many modified bases (7-15/molecule) -
modifications happen to standard bases AFTER
transcription Why the modifications? - may
stabilize 3D structure - may help recognition by
synthetase - Wobble pairing
pseudo uridine
aa
tRNA Secondary Structure
Anticodon loop
8
Figure 27.6
There is significant variability between certain
domains of different tRNAs (2 examples)
E.coli tRNALeu
Mitochondria tRNALys
9
Inosine a modified base also found in tRNA
NH2
guanine has NH2 group
An unusual property of inosine (I) is that it can
base pair with A,C or U - this makes it very
useful in many experimental techniques
(e.g., degenerate PCR where one is uncertain
about the precise sequence the primer should have
at certain locations)
10
The Structure of tRNA
11
Along with unusual / modified bases we also
observe unusual base pairs in tRNA this is due
to the non-regular structure (packing/folding) of
the tRNA compared to DNA, for example
Figure 27.9
12
tRNA Synthetases
Function
- Associates correct amino acid ?? tRNA pair
- Generates a high energy aminoacyl covalent bond
between the two (cleavage of this bond releases
energy and drives protein synthesis) - - Typically one tRNA synthetase exists for each
amino acid and for one or more tRNA
13
General strategy for Synthetasesediting function
- aa binding site excludes amino acids which are
too large and for which there is generally less
affinity - Editing Site is hydrolytic (cleaves)
- - excludes amino acids of the correct size
- - hydrolyzes aa-tRNA linkage of incorrect but
similar amino acids that are small enough to
enter.
Overall error rate 1/40,000
14
tRNA lies across the protein and makes specific
contacts- anticodon region and acceptor stem
3 acceptor stem
ATP
Structure of tRNA sythetase bound to tRNA and ATP
15
Identity Elements in tRNA
- we might predict that the tRNA synthetases
would only use the anticodon to determine the
tRNA identity (bind the correct tRNA)
- what the enzyme actually does is examine other
unique structural features that identify the
tRNA - the location of some of these are
indicated in the secondary structures shown in
the figure
Figure 27.11
16
Formation of aminoacyl tRNA (charged tRNA) by
the aminoacyl tRNA synthetases
Figure 27.10
17
The Genetic Code
How does the aa- charged tRNA read the
mRNA? Since there are 20 amino acids, the
nucleotides must be capable of specifying at
least 20 words (anti-codons) If 2 nucleotides /
amino acid. ? 42 16 possibilities If 3
nucleotides / amino acid. ? 43 64
possibilities
(More than enough)
The actual arrangement of a three letter code has
several possible ways in which it could be
read - overlapping code - punctuated code -
unpunctuated code
18
During protein synthesis, the sequence of
nucleotides in the mRNA is read from the 5 to 3
end in sequential sets of three nucleotides
(codons). In principle, there are three possible
reading frames in protein synthesis however,
only one is correct i.e. codes for the proper
protein. Note that the insertion of one or two
bases will shift the reading frame so that all
amino acids thereafter will be incorrect and the
protein will almost certainly be non-functional.
19
Figure 27.1
not seen
not seen
20
How was the Genetic Code Cracked?
1961 M. Nirenberg used a cell free system to
study the incorporation of 14C-labeled amino
acids into proteins, after the addition of mRNA -
mRNAs ? low level of 14C-phe incorporated -
Poly U RNAs ? HIGH level of 14C-phe
incorporated Poly U encodes Poly Phe UUU ?
Phe AAA ? Lys CCC ? Pro
21
Various kinds of copolymers made Polynucleotide
phosphorylase UDP(75) GDP(25) Random
copolymer containing UG ? 31 -UUU- (3/4)(3/4)(3
/4) 27/64 42 Phe incorporation -UUG- (3/4)(
3/4)(1/4) 9/64 14 Leu -UGU- (3/4)(1/4)(3/4
) 9/64 14 Cys -GUU- (1/4)(3/4)(3/4)
9/64 14 Val -UGG- (3/4)(1/4)(1/4) 3/64
5 Trp ETC..
22
Early 1960s G. Khorana -used synthetic
copolymers of known sequence - triplets and
tetra nucleotide repeats - with this work and the
data of Leder and Nirenberg, the entire genetic
code was completed
Figure 27.2
23
Degeneracy of the Genetic CodeThe Wobble
Hypothesis F.Crick
- - we have 61 codons for 20 amino acids. Each
codon codes for only one amino acid. - Most amino acids have more than one codon!
- Arg, Ser, Leu ? 6 codons
- Gly, Thr, Ala, Val, Pro ? 4 codons
- Met, Trp ? 1 codon
- the code is degenerate- this is important- if
only one codon/ amino acid, most mutational
changes would stop protein synthesis
24
5
5
Ser
Tyr
25
F. Crick proposed that the standard rules for
base-pairing (GC, AU) were relaxed for the
codon at position 3- (see Table 27.2 of Mathews)
aa
Inosine (I) allows types of wobble base
pairs IA IC IU IG GU
3
5
CCI
5-NNNNNGGCNNNNN-3
A
not allowed
U
Flexibility of anti-codon the environment of
the anti-codon in the ribosome both allows
stabilizes wobble pairs
26
Base-pairing possibilities in wobble pairs
Table 27-2 of Mathews et al
27
Structure of the GU Wobble base pair
Figure 27.4
28
The genetic code is almost universal, but there
are a few exceptions / variations Eukaryotic
Mitochondria (human) UGA Trp (not stop) AUA
Met (not Ile) AGA Stop (not Arg) AGG
5
5
Tyr
Ser
Changes in nuclear codes of certain ciliated
protozoans (a class of euks) AGA AGG
STOP
- strong selective pressure to maintain the
genetic code in its current form
29
The Ribosome Structure and Assembly
Large 50S subunit
tRNA (3 bound)
Small 30S subunit
Electron density map of a prokaryotic ribosome
30
Components of the Ribosome
The components of prokaryotic and eukaryotic
ribosomes are essentially identical, although
there are more proteins/RNAs in the eukaryotic
ribosome
31
Secondary structure of 16S ribosomal RNA (rRNA)
from E. coli. - very large complex - various
ribosomal proteins are associated with discrete
areas of the RNA - initially determined by a
variety of biochemical methods
32
Overview of the ribosome structure
33
RNA Protein Ribosome components can be purified
separately reconstituted Such reconstituted
ribosomes are active and SHOW - that all
components are known - they have the capacity for
self-assembly - that mixing and matching
experiments (from diff. organisms) are
possible Detailed structural studies have mapped
the sites of various proteins activities of the
ribosome
34
The Mechanisms of Translation
The process can be separated into 3 different
stages
1. Initiation 2. Elongation 3. Termination - All
stages require mRNA, ribosomes and aa-tRNAs - At
each step there are ALSO other specific protein
factors
35
Table 27.4
36
Initiation
3 key proteins involved, these are known as
Initiation Factors (IFs)
IF 1 3 aid in the disassociation of the 30S
50S subunits. IF 2 - a GTPase which presents
the initiator tRNA
Figure 27.20
37
How does the ribosome know where to begin protein
synthesis?
- Most proteins begin on AUG (GUG UUG less
frequently) - The first amino acid incorporated (in
prokaryotes) is Methionine. - - Methionine of an initiator tRNA is modified
Formylated
O R
(amino acid side chain)
HC-NH-CH-COOH
- Note f-Met looks like a peptide! - normal
tRNAmet only recognizes AUG while tRNAf-Met will
recognize AUG, GUG UUG codons - f-Met only used
at initiation
38
Finding the right AUG or start codon Prokaryotes
Shine-Delgarno sequence on every cistron
(i.e., more than one on polycistronic mRNA
derived from an operon)
Table 27.3
About 5-10 nucleotides upstream of AUG start
codon. Recognized by base pairing to 16S rRNA
(small subunit)
39
Eukaryotes Different - each mRNA is
monocistronic (only one ribosome binding site
needed per mRNA) - greater than 90 of the start
codons used for initiation are those that are
closest to the 5 end - the 5 - CAP, appears to
be recognized by the ribosome
40
Elongation
mRNA is always read 5 ? 3 protein synthesis
is N ? C Involves protein factors known as
elongation factors (EFs) - the 70S initiation
complex is presented with aa-tRNA in the A site
(aminoacyl) by EF-Tu - if correct codon-anticodon
interactions occur, the EF-Tu uses GTP to
completely insert the tRNA - EF-Tu GDP is
released recycled - this is the slowest (rate
determining) step of translation - a balance
between speed and accuracy - error rate 10-4
41
Peptide Bond Formation - the formation of the
peptide bond is catalyzed by the rRNA in the 50S
large subunit - after formation of the peptide
bond the ribosome is translocated or moved down
the RNA - the new tRNA which now has the nascent
peptide is now moved to the P site (peptidyl) -
the tRNA that was previously attached to the
peptide is then moved to the E site (exit), then
released
42
Elongation
Figure 27.22
43
Figure 27.5 - Schematic of the Ribosome
nascent polypeptide chain
44
Termination
- termination requires Release Factors (RFs) GTP
RF1 recognizes UAA and UAG RF2 recognizes UAA
and UGA
Figure 27.26
45
Polyribosomes
-found in both prokaryotes eukaryotes
46
Summary ofProkaryotic vs. EukaryoticTranslation
- general processes are very similar - eukaryotic
ribosomes are larger. 4.2 MDa vs 2.7 MDa - in
Eukaryotes, special MetinitiatortRNA is NOT
formylated - IMPORTANT no euk. Shine-Delgarno
sequence. Initiation occurs at the first start
codon after 5-cap - additional initiation and
Elongation Factors required in eukaryotes
47
- At least two kinds of methods for protein
degradation - The lysosomal system-proteolytic enzymes in
lysosome degrade any trapped protein-not very
selective. - Cytosolic degradation-highly selective
See p1107-1110 of Mathews
48
2) involves tagging with ubiquitin and a
multi-step pathway that usually leads to cleavage
of target protein by multicatalytic proteasome
complex
Figure 28.43 of Mathews