Dispersal models - PowerPoint PPT Presentation
Title:
Dispersal models
Description:
FST, the fixation index, is the most important statistic ... of the mantis schrimp Haptosquilla pulchella. 213 individuals. 106 haplotypes. Minimum-spanning ... – PowerPoint PPT presentation
Number of Views:107
Avg rating:3.0/5.0
Title: Dispersal models
1
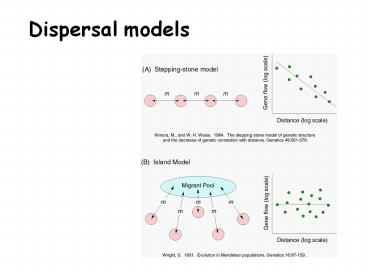
Dispersal models
- Continuous populations
- Isolation-by-distance
- Discrete populations
- Stepping-stone
- Island model
2
FST, the fixation index, is the most important
statistic used to measure the extent of
population subdivision. Where Ht is the
heterozygosity expected to be shown by an
individual in a randomly-mating total
population HS is the mean heterozygosity in
subpopulation Heterozygosity
Where, m is the number of alleles and xi the
frequency of the ith allele at the locus
3
Fine scale genetic structure between populations
of the mantis schrimp Haptosquilla pulchella
Planktonic larvae 4-6 weeks
Dispersal potential estimated at 600 km
Barber et al. 2000
4
New approach in population genetics the
coalescent
5
The coalescent The increasing amount of genetic
polymorphism revealed at the level of DNA
encouraged the use of gene genealogies in
population genetics (the phylogenetic
relationships of the sequences sampled from a
population). Coalescence process The n
sequences in a phylogenetic tree coalesce first
to n-1 ancestral sequences, then to n-2, and so
on until finally to a single common ancestral
sequence, the MRCA most recent common
ancestor.
Under the assumption of genetic drift only, the
probability that 2 alleles share a common
ancestor in the previous generation is 1/2N. If
the population maintained a constant size, the
time back to the MRCA is 4N generations (as we go
backward, there are less lineages left to
coalesce, so that half the coalescence time will
occur between the last 2 lineages).
MRCA
6
Past
7
Coalescence events are more likely to occur in a
small population, or if the allele is subject to
positive selection. The relative distribution of
the coalescent events along the tree, as well as
the total coalescence time, give information
about past and present evolutionary processes in
the analyzed population.
8
How does speciation occur in the ocean ?
9
Molecular Phylogenetics
10
MOLECULAR PHYLOGENETICS The reconstruction of
the evolutionary history between reproductively
isolated species.
The first and crucial point is to compare
orthologous genes.
Then the following steps are required for the
reconstruction of the phylogenetic tree 1)
Alignment of the DNA sequences insertion of
gaps. 2) Choice of the best model of DNA
substitution fitting your DNA alignment. 3)
Inference of the molecular tree using one or
several of the 3 classical methods distance,
maximum parsimony (MP), or maximum likelihood
(ML). 4) Statistical test of the robustness of
the phylogenetic groups (i.e. Bootstrap)
eventually test of alternating phylogenetic
hypotheses (i.e . LRT, likelihood ratio test).
11
DNA sequences Multiple Alignment
12
Assumptions of the model of nucleotide
substitutions
1) All nucleotide sites change independently
2) The substitution rate is constant over time
and in different lineages
3) The base composition is at equilibrium
3) The conditional probabilities of nucleotide
substitutions are the same for all sites and
do not change over time