Translation Overview - PowerPoint PPT Presentation
1 / 17
Title:
Translation Overview
Description:
Termination of Translation ... Following translation by single ribosomes one codon at a time ... Internal Shine-Dalgarno sequences tend to cause translation arrests. ... – PowerPoint PPT presentation
Number of Views:207
Avg rating:3.0/5.0
Title: Translation Overview
1
Translation Overview
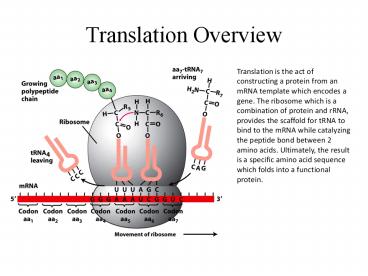
Translation is the act of constructing a protein
from an mRNA template which encodes a gene. The
ribosome which is a combination of protein and
rRNA, provides the scaffold for tRNA to bind to
the mRNA while catalyzing the peptide bond
between 2 amino acids. Ultimately, the result is
a specific amino acid sequence which folds into a
functional protein.
2
Genetic Code
Each tRNA binds to mRNA via an anticodon. An
anticodon is a three nucleotide sequence
complementary to the codon, which is the building
block of a gene. Each codon will bind a specific
tRNA which in turn binds a specific amino acid
and ultimately builds a polypeptide chain. This
table lists which nucleotide sequences encode
which specific amino acid. There are also start
and stop sequences.
3
Initiation of Protein Synthesis
4
Elongation of Polypeptide Chain In Protein
Synthesis
5
Ribosome Structure
Most of the ribosome structure is rRNA with
protein decorations. The A site is for the
aminoacyl tRNA, P for the peptidyl tRNA, and E
for the exiting tRNA. There are two subunits,
which bind at different times in the initiation
step.
6
tRNA Structure
7
tRNA Charging
tRNA charging is the process of attaching the
specific amino acid to its tRNA. The enzyme which
catalyzes this reaction is aminoacyl tRNA
synthetase. Like the tRNAs themselves, the
synthetases are specific to amino acids, and thus
the tRNAs as well.
8
Termination of Translation
The termination of translation begins when a stop
codon translates into the A site of a ribosome.
Some kind of release factor then moves into the A
site of the ribosome (The picture on the left is
for eukaryotic translation termination). The
release factor is a protein which is bound to a
triphosphorylated nucleotide whose hydrolyzation
facilitates the disassembly of the
ribosome-tRNA-peptide chain complex.
9
Shine-Dalgarno Sequences
A Shine-Dalgarno sequence is the ribosomal
binding site of a ribosome in a prokaryote. What
is shown in the figure above is the rRNA in the
16s subunit of a prokaryotic ribosome whose
nucleotide sequence is complementary to the S-D
sequence, called the anti Shine-Dalgarno
sequence. It is important to note that a
Shine-Dalgarno sequence internally found in a
prokaryotic gene can render the gene
polycistronic, i.e. another ribosome can bind to
the internal S-D sequence and begin translation
from a different point in the gene if a start
codon (AUG) is in relatively close proximity.
This can lead to genetic frameshifting and
protein misfolding.
10
Following translation by single ribosomes one
codon at a timeWen, Jin-Der Lancaster, Laura
Hodges, Courtney Zeri, Ana-Carolina Yoshimura,
Shige H. Noller, Harry F. Bustamante, Carlos
Tinoco, Ignacio
- Michael Boyle
11
The experimental setup of the paper is shown in
A. The experimental hairpin mRNA/Ribosome complex
is held in place between two polystyrene beads
held in place with an optical tweezer and
micropipette respectively. B shows the primary
nucleotide structure of the hairpin mRNA. The
Shine-Dalgarno and start codon are highlighted in
blue, the places where the ribosome is to be
stalled are boxed, and the valine and glutamic
acid patches are highlighted in gray and yellow,
respectively.
12
Control Experiments
- To verify that the change in distance was due to
mechanical unfolding of the hairpin by a ribosome - Used a known mRNA construct that encoded certain
amino acids. The mRNA contained a codon at a
desired stall location that encoded an amino acid
which was not added into the translation mixture.
When extension was stalled, puromycin was added
to the mixture and the mRNA refolded into its
hairpin structure shortly after, confirming the
first bullet.
13
Optical Tweezers
An optical tweezer is a method in which to trap a
micron or nano-sized particle in a fixed
location. It works by shining a focused laser
beam. The center of the beam, called the waist is
the thinnest and has the highest electric charge
gradient. Charged particles are attracted to the
waist of the laser beam however they are trapped
a finite distance upstream from the waist beam
because of the force the laser beam exhibits on
the particle in the direction that the laser is
propagated.
14
The figure shows the step-pause-step pattern of
the ribosome translating the VE60hp . The
plateaus represent the pause time in between
translocation events, i.e. translation. The steep
sloped areas in between translation represent the
unwinding of the mRNA hairpin and the
translocation of the ribosome to the next codon,
which occur within 0.078 s of each other. The
translocation times are then grouped into 0.025 s
bins and fit with different mechanism models, of
which the three step mechanism fit best.
15
The first graph shows the number of bases
translocated as a function of time for 4
different ribosomes. Some of the ribosomes pause
for long times and translation even arrests for
some of the ribosomes. b.) shows a closeup look
of the ribosome trajectories.
16
A.) shows the distribution of dwell times for the
VE274hp mRNA construct fitted with a curve that
corresponds to a two step mechanism, which they
determined to be best for all ribosomes and dwell
time subsets for the VE274hp mRNA. B.) shows the
trend that the dwell time increases as the force
of the optical tweezer decreases. This is due to
the tension that the tweezer has on the
experimental setup, either increasing or
decreasing the energy needed to open the mRNA
hairpin. C.) Shows the effect of force on long
pauses. As the force goes down, the ribosome goes
into a long pause. Once the force is rapidly
increased, the ribosome continues translating
immediately.
17
Biological Significance/Conclusions
- Follows ribosomes in real time and reveals that
translation is a step-pause-step process, not
continuous - Gives a time frame for translation- 2.2 s median
for pause time and - These time frames can now be examined for bulk
translation experiments in order to better
correlate biochemical results and come up with a
kinetic model for translation - Internal Shine-Dalgarno sequences tend to cause
translation arrests. These arrests could lead to
translational frameshifting and protein
misfolding. - Future work should look into tethering a single
ribosome so as to determine its motor properties




























![❤️[READ]✔️ The Routledge Handbook of Translation, Interpreting and Crisis (Routledge Handbooks i PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10053360.th0.jpg?_=20240611113)
![[Read] The Routledge Handbook of Translation, Interpreting and Crisis (Routledge Handbooks i PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10055285.th0.jpg?_=20240614114)
