Multifunctional Protein Microarrays - PowerPoint PPT Presentation
1 / 40
Title:
Multifunctional Protein Microarrays
Description:
Clones code for a tagged protein (GST, FLAG, HA...). When an anti-tag ... Challenges of Cloning. B.anthracis Genome. AT richness of ORFeome. optimize PCR condition ... – PowerPoint PPT presentation
Number of Views:301
Avg rating:3.0/5.0
Title: Multifunctional Protein Microarrays
1
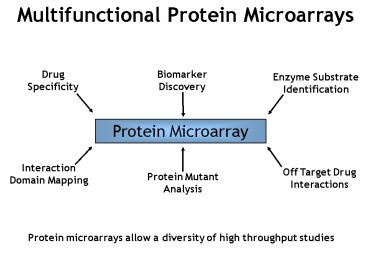
Multifunctional Protein Microarrays
Protein microarrays allow a diversity of high
throughput studies
2
NAPPA Nucleic Acid Programmable Protein Array
Easy to make DNA arrays are converted to ready to
use protein arrays
3
NAPPA rationale
4
Specific proteins are produced in situ
Specific anti-Prot D detection
spotted clone
spotted clone
Prot D
Clones code for a tagged protein (GST, FLAG,
HA). When an anti-tag antibody is used for
detection all proteins light up. Specific
antibodies show proteins are correctly captured
in situ after expression
5
Protein expression on NAPPA
Over 10,000 genes expressed
- Average protein yield 20 fmol per feature
- Approx. 1000 fold more than protein spotting
arrays - 95 of proteins express and capture well on
NAPPA - 10, 000 proteins tested
- Multiple organisms tested
- Membrane proteins express well
6
Combining NAPPA with serum screening to find
immunodominant antigens
7
Biomarker Discovery
Validation and Discovery of informative antigens
in One Step
Replicate microscopic arrays of candidate antigens
NAPPA microarrays can be used for serum screening
as any other Protein microarray, with the
advantage that proteins are produced just in the
moment of experimentation, avoiding
degradation and increasing shelf life of slides
8
Identifying immunodominant antigens in infectious
diseases
High density display of pathogen proteomes
Pseudomonas aeruginosa (300 select
genes)
Francisella tularensis proteome (1700 genes)
Vibrio cholerae proteome (4000 genes)
9
Serum screening of a series of cystic fibrosis
patients infected with Pseudomonas aeruginosa
Patient 1
Patient 2
Patient 3
Control 1
Control 2
Patient 6
Patient 5
Patient 4
Pseudomonas aeruginosa (300 select
genes)
In this case, positive spots can be identified
even by visual observation
10
Preliminary results from Vibrio cholerae serum
screening
PATIENT A
PATIENT B
acute
convalescent
acute
convalescent
partial slide map
EBNA
tcpA
EBNA
tcpA
In the acute phase patients respond only to the
positive control EBNA (Epstein Barr viral
protein) In the convalescent phase they also
respond to Vibrio cholerae proteins as tcpA
11
Vibrio cholerae serum screening
PATIENT 372
PATIENT 8
CONVALESCENT
CONVALESCENT
ACUTE
ACUTE
PATIENT 375
PATIENT 373
CONVALESCENT
CONVALESCENT
ACUTE
ACUTE
Acute x Convalescent plots for the same patient
show hits in convalescent phase
12
Comparison of NAPPA and traditional ELISA
technique
Results are pretty comparable
13
Other possible NAPPA applications
14
Protein-protein interaction
Mapping the Human DNA Replication complex
15
Interaction Map
- 1000 potential interactions interrogated
- 110 interactions observed
- 85 of all known binary interactions were
detected - 65 novel interactions identified
- All interactions exhibited expected selectivity
16
Protein small molecules interaction studies
17
Clone Production and Validation
Production of V.cholerae and B.anthracis ORFeome
Collections
18
Public databases
Public databases
Import ORF information
Validate clone sequence
Submit clone info to Genbank
Clone Production
FLEXGene
ACE
Send clones to distributors (eg. PFGRC)
Under construction data tracking systems
NAPPABase CellBase
PlasmID
shRNA clones
Use clones for proteomics studies
Distribute clones through DF/HCC
cDNA clones from other sources
19
Gel Purify
Clone
Transform
Sequence Verify
20
ORF Configuration
- Normalized initiation and omit stop codons
Kozak
21
Vibrio cholerae ORF Collection
- Clone Production and Validation
22
V.cholerae Genome Information
ORF information N16961 strain, chromosome 1 and
2 TIGR information supplemented with NCBI
GC content AVG47.0
ORF count 3888
23
Clone ProductionWorkflow DiagramPart 1 of 2
Continues with Capture Reaction
24
Clone ProductionWorkflow DiagramPart 2 of 2
After gel/PCR
25
Sequence ValidationWorkflowDiagram
26
Sequence Validation of V.cholerae FLEXGene
ClonesProject Summary
27
Quality of V.choleraeFLEXGene Collection
Finished ORFs 3760 (97) --- total of
V.cholerae genome 3503 (93) -- perfect
match 77 (2) -- silent 154
(4.1) -- 1 missense 26 (0.7) -- ins/del
(correct authentic frameshift)
28
Bacillus anthracis ORF Collection
- Clone Production and Validation
29
B.anthracis Genome Information
ORF information Ames strain, chromosome/pXO2
from NCBI
GC content AVG35.4
ORF count 5415
30
Challenges of Cloning B.anthracis Genome
- AT richness of ORFeome
- optimize PCR condition
- more cycles than usual due to inefficient
amplification - Difficulties to obtain genomic DNA
- Size of genome
31
Set-up for Cloning
32
Cloning Workflow and Decision Points (clones from
genomic DNA)
33
(No Transcript)
34
Preparation of Clones for Sequencing
35
DNA sequencing and analysis
36
Detailed cloning schema part 1
37
Detailed cloning schema part 2
38
Sequence Validation of B. anthracis FLEXGene
ClonesProject Summary
39
Quality of B.anthracis FLEXGene Collection
Finished ORFs 5153 (92.4) --- total of B.
anthracis genome 4720 (91.8) -- no amino acid
change 359 (7.2) -- 1 missense
40
Progress in Building FLEX
End Reads
Full Length
Cloning
- S. cerevisiae 6,000 genes Gateway
- H. sapiens 4,000 genes (x2) Gateway
- 3,500 genes(x2) Creator
- P. aeruginosa 5,570 genes Gateway
- Y. pestis 4,200 genes Gateway
- F. tularensis 1,603 genes Gateway
- V. cholerae 3,888 genes Gateway
- B. anthracis 5,415 genes Gateway
- Dengue WNV 20 genes Gateway
- Falciparum 257 genes Gateway
- P. Pseudotuberculosis 306 genes Gateway
?
?
?
?
?
?
?
1/2
?
?
?
?
?
?
?
?
?
?
?
41
HIP Clone Overview