Introduction: Logic of Gene Regulation - PowerPoint PPT Presentation
1 / 1
Title:
Introduction: Logic of Gene Regulation
Description:
Gene regulation takes place when a transcription factor proteins produced by one ... Brock et al have reported the limiting behavior of fuzzy logic model as the ... – PowerPoint PPT presentation
Number of Views:48
Avg rating:3.0/5.0
Title: Introduction: Logic of Gene Regulation
1
Fuzzy Model for Gene Regulatory NetworkRamesh
Ram, Madhu Chetty and Trevor I. DixGippsland
School of IT, Northways Road, Churchill, VIC
3842, AUSTRALIAramesh.ram, madhu.chetty,
trevor.dix _at_ infotech.monash.edu.au
Abstract Gene regulatory networks influence
development and evolution in living organism. The
advent of microarray technology has challenged
computer scientists to develop better algorithms
for modeling the underlying regulatory
relationship in between the genes. Here, we
present a fuzzy logic model for the detection of
activator-repressor regulatory networks from
microarray data. In addition, we introduce a
novel pre-processing technique that eliminates
redundant computation performed by the proposed
model, hence optimizing the computation time.
Saccharomyces cerevisiae microarray data was
applied to the model and 548 activator/repressor
regulatory triplets were inferred from the model.
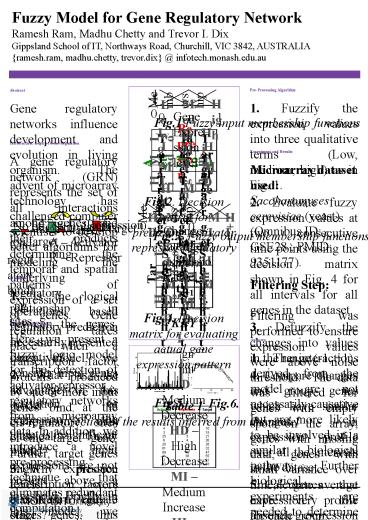
Pre-Processing Algorithm 1. Fuzzify the
expression values into three qualitative terms
(Low, medium, high) as in Fig. 1. 2. Evaluate
fuzzy expression values at two consecutive time
points using the decision matrix shown in Fig. 4
for all intervals for all genes in the
dataset. 3. Defuzzify the changes into values in
the range (-1, 1) 4. Compare changes and group
genes which have similar changes in expression
profile over all the intervals in the dataset.
5. Calculate average expression profile for
each group.
Fig.1 Fuzzy input membership functions
Repressor
Low
Medium
High
Target
Target
Target
Low
I
MD
HD
Introduction Logic of Gene Regulation A gene
regulatory network (GRN) represents the set of
all interactions among genes and their products
determining the temporal and spatial patterns of
expression of a set of genes. Gene regulation
takes place when a transcription factor proteins
produced by one or more input genes bind at the
cis-regulatory sites of the target gene. Further,
target genes might produce transcription factors
that in turn regulate other genes, thus forming a
complex network. Expression of target gene is
dependent on the expression of genes producing
its transcription factors. The biochemistry of
gene expression relates transcription to
combinatorial logic of the input gene
expressions. Most commonly found logic in
biology is the activator-repressor logic. A gene
can be called activator, If expression of output
gene increases with increase in expression of
input gene, while a gene is called repressor if
expression of output gene decreases with increase
in expression of input gene. The figure below
shows the activator-repressor logic. Green
indicates expression of activator gene while red
indicates expression of repressor gene. At high
and low expression levels of the input, the
target expression can be easily stated as shown
above. Yet, gene expression is continuous process
rather than discrete, hence we use fuzzy values
to bridge the gap and predict regulatory
relationship.
Experiment and Results Microarray dataset used
Saccharomyces cerevisiae (yeast) (Omnibus ID
GSE28 PMID 9351177). Filtering Step
Filtering was performed to ensure expression
values were above noise threshold. The data was
filtered for genes with empty spots on the array,
genes with missing data, genes with small
variance over time, genes that have very low
absolute expression values and genes whose
profiles have low entropy. After filtering 310
genes remained out of 6321 gene data.
Normalization Step Min-Max Technique Preprocessi
ng Step The presented
preprocessing algorithm was applied on the data.
There were 101 unique patterns of changes in
expression profile observed in the dataset of 310
genes. Fig. 5 shows the bar plot of the frequency
of occurrence of these 101 patterns. Out of 310
patterns, 58 patterns were found to be present
independently and the remaining patterns formed
43 groups due to similarity in their changes in
expression profile. An average expression profile
for each group was computed and was used as
reference input to the fuzzy GRN model. As an
example, in Fig. 6 the set of genes belonging to
the 42nd pattern of expression profile is grouped
using the pre-processing algorithm. Model
Evaluation The expression profiles corresponding
to these 101 patterns were fed as input to the
fuzzy model. There were a total of 5050
(n(n-1)/2) possible combinations of activator/
repressor pairs as inputs to the model. The
inputs were fed to the fuzzy model and evaluated
using the decision matrix shown in Fig. 1. The
5050 predicted patterns were compared against the
precomputed actual patterns of changes in
expression profile excluding the input patterns.
Out of 5050 combinations analysed, 548
combinations of predicted output were validated
and are likely to have activator/ repressor
regulatory relationships. Results Summary The
coding was done in Matlab. Table 1 shows some of
the results inferred using the fuzzy logic model.
The numbers correspond to expression profiles
derived from the data. Remaining results are
available with the authors. The computation time
was relatively small. Hence our method will be
able handle larger datasets at lower computation
time. The model can be extended to implement
other combinational logical operations such as
AND, OR and so on by modifying the corresponding
decision matrix.
Target
Target
Target
Medium
Activator
MI
I
MD
Target
Target
Target
High
I
MI
HI
Fig.2. Decision matrix for predicting Activator
repressor regulatory pattern
1
I
MD
HD
MI
HI
Membership
0
0
-1
1
Change in target expression level
Fig.3. Fuzzy output membership functions
Target at tn1
Low
Medium
High
Shift
Shift
Shift
Low
MI
HI
I
Shift
Shift
Shift
Target at tn
Medium
Questions 1. Is the logical operations based
regulatory prediction necessarily causative? 2.
What is the advantage of this model in comparison
with classical Fuzzy model? 3. Why expression
levels were classified to only 3 stages? 4. How
does the proposed pre-processing technique differ
from clustering techniques?
MD
I
MI
Shift
Shift
Shift
I
MD
HD
High
n is number of time points in the data
Fig.4. Decision matrix for evaluating actual gene
expression pattern
KEY I Insignificant MD Medium Decrease HD
High Decrease MI Medium Increase HI High
Increase
Presented Fuzzy Model In the presented fuzzy
model, we consider input genes as drivers, i.e.
activation or repression of the target is
insignificant when input expressions are not
present above a threshold. Briefly, in this
model, we predict changes in expression of the
target gene over interval time points based on
input expression, and compare the predicted
pattern with actual changes in expression of all
remaining genes. The model works as follows. 1.
The expression values of all genes are normalized
to the range 0 and This is done using min-max
technique 2. Expression values are then
classified using three fuzzy membership functions
namely low, medium and high as in Fig. 1. 3. All
combination of two input genes are selected from
the data and evaluated using the heuristic fuzzy
rules for activator/repressor regulatory logic
(as in Fig. 2.) 4. The output of the system has
five fuzzy membership functions as shown in Fig.
3 describing various possible changes in
expression. The result is a set of predicted
patterns of changes in gene expression over
entire interval time points present in the data
for all combination of pairs of
activator-repressor input genes. 5. Now, each
fuzzified gene expression profile is evaluated
using the heuristic rules for finding actual
changes in expression (shown in Fig. 4). The
result is a set of patterns of actual changes in
gene expression over entire interval time points
given in the data. 6. The predicted and actual
patterns are compared and the genes whose
expression pattern matches with the predicted
pattern are taken as target genes to the
corresponding input genes. This results in
triplets of Activator-repressor and target genes.
These regulatory triplets best fit the model for
the given data. There is a possibility that
predicted changes in expression pattern can match
to two or more actual expression pattern. This is
due to presence of similar expression profiles in
the data. This difficulty is solved by using the
proposed novel preprocessing technique explained
next.
Answers 1. The interactions derived from the
model are not necessarily causative but are more
likely to be involved in a similar biological
pathway. Further biological experiments are
needed to determine the validity of the genetic
interactions suggested by the model. 2. The
fuzzy decision matrix used in the classical model
is capable of giving possible false predictions.
For instance, consider the input activator and
repressor expressions to be classified Low at
time t by the fuzzy model, i.e. the input genes
are considered to have no effect on the target
gene expression in the interval time t and t1.
The classical fuzzy model predicts the output
expression level as medium which is true in
most cases. However, in a scenario where the
target expression level was low at time t-1,
the classical model is actually predicting the
target expression to increase from Low to
Medium level when the activator is low. This
would result in a false prediction as the target
expression can be predicted to increase
significantly only when the activator is above
threshold expression level. For the above
scenario, the proposed model predicts an
insignificant change with respect to the inputs
and thus prevents a false prediction. 3. Brock
et al have reported the limiting behavior of
fuzzy logic model as the number of classification
states tends toward infinity and the classical
fuzzy model using three state classification has
produced biologically plausible results. 4.
The presented pre-processing algorithm groups
genes which have similar changes in their
expression profiles (in other words similar fuzzy
values). Compared to clustering techniques, the
method does not require specification of a number
of clusters and all useful information in the
dataset is taken for analysis. In addition, the
process eliminates redundant computation from the
model and optimizes computation time.
Fig.5.
Fig.6.
Table 1 Section of the results inferred from the
model