Physical Properties - PowerPoint PPT Presentation
1 / 225
Title:
Physical Properties
Description:
Hydrogen bond- weak attraction between hydrogen on adjacent molecules such as water ... Turgid- Plant cell gains. Plasmolysis. Membrane shrinks due to water loss ... – PowerPoint PPT presentation
Number of Views:144
Avg rating:3.0/5.0
Title: Physical Properties
1
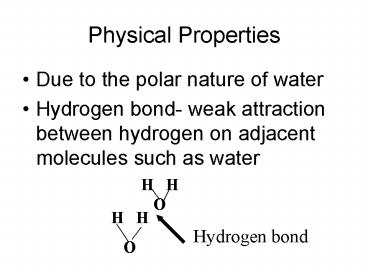
Physical Properties
- Due to the polar nature of water
- Hydrogen bond- weak attraction between hydrogen
on adjacent molecules such as water
H H O
H H O
Hydrogen bond
2
Water and its importance to Life
- Life evolved in water
- Waters unique properties have made life as we
know it possible
3
Physical Properties
- Heat of vaporization- amount of energy that is
released or gained when changing state from
liquid to gas or back
4
Physical Properties
- High Specific Heat- the amount of heat absorbed
or released when water changes temperature by one
degree C. ( 1 cal. )
5
Ice Floats
- As a liquid waters hydrogen bonds continuously
break and reform - As a solid four molecules form hydrogen bonds
creating crystals with open channels and thus
fewer molecules per area.
6
Physical Properties
- Water reaches maximum density at 4 degrees C.
- Water is a universal solvent due to its polar
nature
7
Evaluate the importance of the following and
explain the property of water responsible.
- Cytoplasm is 98 water
- Ice Floats
- Lake effect temperature moderation
- Evaporative Cooling
- Spring-Fall Overturn
8
Most Abundant Chemicals in Life
- Carbon
- Oxygen
- Hydrogen
- Nitrogen
- Ca, P, K, S, Na, Cl, Mg gt 4
96
9
Carbon is special
- Tetrahedral structure- four valence electrons
shared - Covalent bonds - stability
10
Carbon is Special
- Variations are possible in carbon molecules that
provide diversity - Isomers are possible
- structural- differ in structure same
chemical formula - geometric-differ in spatial
relationship - enantiomers-mirror images of each
other
11
Condensation Synthesis
A
B
A
B
H2O
A and B could be monosaccharides or amino acids
12
Hydrolysis
H2O
Addition of water breaks the bond
13
Polymers
Polymers are repeating units of monomers. They
are very important to Biology. They are made or
synthesized by the removal of water called
CONDENSATION SYNTHESIS They are broken down by
the addition of water or HYDROLYSIS
14
Classes of Biomolecules
- Carbohydrates- used for energy and structures(
building living organisms) - Lipids- used for energy storage, communication
and structures - Proteins- used for a variety of life functions
- Nucleic Acids-the instructions for building life
15
Carbohydrates
- Three common forms
- Monosaccharides
- Disaccharides
- Polysaccharides
16
Carbohydrates
- Monosaccharides- single sugars or simple
sugars,ex. Glucose ( C6H12O6) - Disaccharides- double sugar, ex. Sucrose
- Polysaccharides- polymers of glucose such as 1.
Starch 2. Cellulose 3. Glycogen 4. Chitin
17
Review
- What will happen here?
- AOH HB ?
- And here
CH2OH
CH2OH
O
O
OH
OH
H
OH
OH
H
OH
OH
OH
OH
H2O
18
Dehydration Synthesisor a Condensation Reaction
- A B AB H2O
CH2OH
CH2OH
O
O
OH
O
OH
OH
OH
H2O
OH
OH
19
Review
- What will happen here?
- AB H2O ?
- And here
CH2OH
CH2OH
H2O
O
O
OH
O
OH
OH
OH
OH
OH
20
Hydrolysis or Reaction
- AB H2O AOH HB
Molecules have been HYDROLIZED!
CH2OH
CH2OH
O
O
OH
OH
OH
OH
OH
OH
OH
OH
21
Glucose
CH2OH
Glucose has a chemical formula of C6H12O6
C
O
H
OH
OH
C
C
H
OH
H
H
C
C
OH
H
22
FRUCTOSE
O
CH2OH
CH2OH
C
C
H
H
HO
H
C
C
OH
OH
23
Disaccharides
- Sucrose and Lactose
- 2 monosaccharides bonded together
Alpha or Beta?
CH2OH
CH2OH
O
O
O
OH
OH
OH
OH
OH
OH
24
Polysaccharides
- 3 or more Monosaccharides bonded together
CH2OH
CH2OH
CH2OH
O
O
OH
O
O
OH
OH
OH
O
OH
OH
OH
OH
25
Polysaccharide
- Starch-storage in plants
- Cellulose-structural part of plant cell wall
- Glycogen- storage in animals, liver
- Chitin structural component for arthropods,
exoskeleton. Also found in fungi.
26
Polysaccharides Starch
- Plants use it as energy storage
- Difficult for humans to break down
- Ex. Avoid a high starch diet
27
polysaccharides
Glucose monomers
28
Polysaccharides Cellulose(B 1, 4 linkage)
- Long fibers
- Up to 15,000 Glucose units per strand
- Most abundant biological substance on earth
- Ex. Cotton, Trees, Paper
- Why is cellulose so strong?
- Why cant humans breakdown cellulose and cows
can?
29
Polysaccharides Glycogen
- Animals use it as energy storage
- Lots and lots of it in the liver
- Forms huge branched storage units which allow for
easy break down for energy
30
Other polysaccharides
- Chitin
- Found in the exoskeleton of insects, and
arthropods - Ex. Crabs, lobsters, grasshoppers
- Pectin
- Found in plant cell walls
- Provides rigidity
- Heteropolymers
- Glycoproteins and peptidoglycans
31
Protein
Polymers of amino acids With 20 natural amino
acids there are a variety of proteins
32
Amino AcidsThe building blocks of protein
H
O
H
N -C - C
H
OH
R
R- there are twenty different R groups possible
33
Alanine NH2-CH-COOH
Glycine NH2-CH2-COOH
CH3
34
Peptide bond- is a bond between amino acids a
molecule of water is removed
35
Protein Structure
1. Primary- order of the amino acids 2.
Secondary- hydrogen bonds cause pleats and
helix 3. Tertiary- folds and loops create shape
by R Group bonds 4. Quaternary-interaction
of several proteins
36
A protein with secondary structure
37
A protein with Tertiary Structure
38
Lipids
- Large molecules that do NOT have an affinity for
water not soluble in. - May have hydrophobic-water fearing and
hydrophilic-water loving parts.
39
Triglycerides
hydrophilic
hydrophobic
40
Types of Lipids
- Made of hydrocarbons -
- Triglycerides- fats, waxes, and oils(saturated
all single bonds C-C, unsaturated have double CC
bonds - Phospholipids- attached phosphate replaces one
of the hydrocarbon tails - Steroids- Ring Forms of Hydrocarbons cholesterol
and some hormones
41
Triglycerides
- Saturated fats- single bonds make this a solid at
room temperature and more difficult to digest.
42
Unsatured Fats
- Triglycerides that contain double bonds (
dehydrogenated) are liquids at room temp and more
digestable
43
(No Transcript)
44
(No Transcript)
45
(No Transcript)
46
Nucleic Acids
- Made of monomers called nucleotides
- DNA- deoxyribonucleicacid
- RNA- ribonucleic acid
- These molecules carry all the hereditary
information of living things
47
DNA Basic Composition
- DNA is made up of nucleotides
- Nucleotides are made of
- ...Deoxyribose sugar
- Phosphate
- Base
- bases are guanine,cytosine, thymine and adenine
48
RESPIRATION
SYNTHESIS
C A T A B O L I S M
A N A B O L I S M
ATP SYNTHESIS FROM ADP Pi
49
Free Energy
- Ability to do work in the cell or ecosystem.
50
Energy Transfer
- ATP formation
- G
- ENDERGONIC
- Stores energy in phosphate bond
- ATP breakdown
- - G
- EXERGONIC
- Releases energy between phosphates
51
Enzyme Characteristics
- Lower the activation energy
- Speed up the rate of a reaction
- Act as catalysts
- Are proteins (occasionally RNA)
52
Enzyme Characteristics
- Conformation or shape is most important feature (
Lock and Key Hypothesis) - Substrate Specific
- Do NOT become part of reaction
53
(No Transcript)
54
(No Transcript)
55
(No Transcript)
56
(No Transcript)
57
Enzyme activity.
7
pH
58
Enzyme activity.
7
pH
59
(No Transcript)
60
Enzyme activity.
10
Temperature o C
61
Enzyme activity.
30
10
Temperature o C
62
Active site
Allosteric site
63
substrate
Enzyme
products
64
Cofactors
- Non protein helpers for enzyme activity
- May bind to active site tightly or loosely
- Many are inorganic such as zinc or iron
- If organic they are called Coenzymes
65
Allosteric site
- Regulatory site other than the active site.
66
Competitive Inhibitor
substrate
inhibitor
Enzyme-Substrate Complex
Enzyme
Noncompetitive Inhibitor
67
Allosteric Regulation
Active site
activator
Inactive form
Active conformation
Allosteric site
inhibitor
68
Feedback inhibition
- Product may cause negative feedback (act to
inhibit, disrupt conformation) - Reactants may cause positive feedback ( act to
preserve enzyme conformation)
69
Enzyme c
Initial substance
Enzyme b
Enzyme a
End product
- feedback
70
Prokaryotic Cells
- Lack a nucleus
- Lack membrane bound organells
- Include bacteria and other Monerans
71
Eukaryotic Cells
- Have a nucleus
- Have membrane bound organells
- Plants, Animals, Fungi, and Protists have these
cell types
72
(No Transcript)
73
Organells Membrane Bound ( endomembrane) Nucleus
Endoplasmic reticulum (rough) Endoplasmic
reticulum ( smooth ) Golgi apparatus Lysosome Vacu
oles Vesicles Peroxisome ( single membrane)
Mitochoindria Chloroplasts
74
Non membrane bound organells Nucleolus Microtubule
s Microfilaments Centrioles Cilia Flagella
75
Nucleus
- Chromatin- DNA organized with protein (histone)
- Controls Protein Synthesis
- Double Membrane with pores
- may be continious with ER
- Nucleolus- made of and synthesizes RNA
76
(No Transcript)
77
Endoplasmic Reticulum
- Rough ER- contains ribosome for protein synthesis
- Smooth ER- lacks ribosomes, synthesis of lipids,
metabolism of carbohydrates, detoxification of
drugs and poisons - Muscle ER- calcium ion transfer
78
ER and protein synthesis and Transport Vesicles
- Export Proteins become enclsed in vesicle of
the ER Pinch closed - Especially secretory proteins ( glycoproteins )
E R..
79
Golgi Apparatus
- Manufacture, storage shipping , and packaging
secretion products - Cis
- Trans
- Vesicles
trans
cis
80
(No Transcript)
81
Phospholipids are amphipathic - have both
hydrophobic and hydrophilic portions
hydrophilic
hydrophobic
82
(No Transcript)
83
(No Transcript)
84
Membrane Fluidity
- Unsaturated hydrocarbons in the phospholipids
make it flow laterally - Cholesterol maintains some rigidity at low
temperatures and prevents too much fluidity at
high
85
Membrane proteins
- Integral-penetrate into or through the lipid
bilayer - used for transport
- Peripheral protein- attach to the surface of the
lipid layer - used for- receptors, recognition(carbohydrates
attached)
86
(No Transcript)
87
Permeablility
- What passes through easily
- Oxygen
- Carbon dioxide
- water
- What does not pass through easily
- ions
- proteins
- carbohydrates
88
Transport
- Passive
- Requires no expenditure of ATP
- Moves from high to low
- Can be facilitative-aided by protein conformation
change - Gatted channels
- Active
- Requires ATP energy
- Generally moves from low to high
- Gatted channels
- Na-K pump
- Proton Pumps
- Phagocytosis or Pinocytosis
89
Solutions
- Homogeneous-same throughout
- solvent- what you are dissolving into
- solute- what you dissolve
90
solution
SOLUTE
SOLVENT
91
Hypertonic
92
.5M glucose Distilled water 1.5 M glucose
.5M glucose
93
Water Balance
- Plasmolysis( plasmolyzed) plant cell shrinks or
looses water - Flacid -plant cell gains water and looses at same
rate - Turgid- Plant cell gains
94
Plasmolysis
- Membrane shrinks due to water loss
- Restricted to cells with walls
- Occurs in a Hypertonic environment
95
Facilitated Diffusion
- Proteins make movement of polar molecules, ions,
or larger compounds possible by providing a
passage. - Often protein changes conformation
- NO ATP required
- Movement from high to low concentration
96
Active Transport
- Sodium Potasium Pump
- Proton pumps
97
Membrane Potential
- Voltage across a membrane
- -50 to-200 millivolts
- Electrochemical gradients- combination of ion
potential and electric charge difference
98
Gated Channels
- Chemical or electrical impulses cause them to
change shape-OPEN
99
(No Transcript)
100
(No Transcript)
101
Na-K Pump
- Membrane Potential-voltage difference across a
membrane - Chemical Gradient-difference in concentration of
solute across a membrane - Electro-chemical Gradient- combination of the
above ex. - Na,K, Cl-(8.13)
102
Exocytosis and Endocytosis
- Phagocytosis- engulf pseudopodia
- Pinocytosis- gulps
- Receptor mediated pinocytosis
103
Signal Transduction
- Binding of extracellular molecule to receptor
protein see model on pg. 156 Campbell
104
Cell types determine cycle
- Prokaryotes- binary fission circular
chromosome attaches to inner membrane.
Replication is followed by reattachment at two
sites.
- Eukaryotes-have a larger genome and nuclear
genetic material must be carried on several
chromosomes by specialized structures.
No spindle fibers
105
Can be Confusing?
Chomosomes Chromatids Sister chromatids Homologous
chromosomes Centromere Centrosome Centrioles Kine
tochore Nonkinetochore
106
Chromosomes
DNA is continuous and wound around protein which
is coiled and super coiled into a dark staining
body. Chromosomes can be seen as having two arms
and often one is longer. When duplicated the
chromosome has identical sister chromatids held
together at the centromere.
107
chromatid
centromere
Sister chromatid
108
Chromosome Number is fixed in each species
109
Cell Cycle
Events in the growth, development
and reproduction of the cell. Go cells have
stopped dividing or have lost the potential to
divide.
110
(No Transcript)
111
G1- gap or growth after cell division. Cell
grows in size. this stage contains the
RESTRICTION point S- synthesis of new DNA from
existing template(replication) G2- gap 2 or
growth prior to cell division M- mitosis or
chromosome division C- cytokinesis or cell
division
Interphase G1, S, and G2
112
Control of Cell Cycle
Restriction point- go/no go control during G1 G0
- a non-dividing stage for a cell Growth
Factors-compounds which regulate growth and
division. Ex.PDGF platelet derived growth
factor Density-dependent inhibition- crowding
inhibits cell division. Adhesiveness- cells ECM
causes them to stick together Metastasis-cells(ca
ncerous) migrate
113
Cell Clock Regulators
- Proteins ( enzymes) regulate cell cycle
- Produced by internal cell clock genes
- Protooncogenes- cause cells to divide
- Tumor suppressor genes- prevent cell division
114
Cancer and the Cell Cycle
- Normal Cells
- Adhesive
- Contact inhibition
- Cancer cells
- Lost adhesiveness
- Lost contact inhibition
115
(No Transcript)
116
Principles of Heredity
- Alternative versions of genes (alleles) account
for variations in a trait. - For each character, an organism inherits two
alleles, one from each parent. - If alleles differ, then the dominant will be
fully expressed over the recessive. - The two alleles segregate (separate) during
gamete formation. - Alleles on different chromosomes segregate
independently of one another
117
Crossing over
- During prophase of meiosis homologous pairs may
exchange genetic material.
118
(No Transcript)
119
New Genetic Combinations
- Recombination during fertilization brings
together two sets of genetic instructions - Meiosis-crossing over brings about new
combinations - Random genetic mutation can result in random
genetic change
120
Electron Carriers
- NAD nicotinamide adenine dinucleotide
- NAD When oxidized
- NADH 2 H When reduced
- FAD or FADH2
121
Types of Respiration
- Anaerobic-without oxygen 1. Alcoholic fermentaion
2.Acetic Acid fermentaion 3. Lactic Acid
fermentaion - Aerobic-with oxygen
- ALL OF THESE BEGIN WITH THE ANAEROBIC PROCESS OF
GLYCOLYSIS
122
GLYCOLYSIS
Glucose is made ready to metabolize by addition
of phosphates and then it is broken down into
2- 3 carbon compounds (PYRUVATE)
This yields a net gain of 2 ATP
123
(No Transcript)
124
ACETYL COA FRORMATION
- Pyruvate is converted into a 2 Carbon compound
and added to an enzyme - CO2 is released
125
Krebs Cycle
- Breaks C-H-O bonds
- Energy is transferred via carriers to other steps
- CO2 released
- Some small amount of ATP is produced
126
Electron Transport
- Hydrogen Pathway- pumps H ions
- Electron Transfer
- Chemiosmosis- H ions flow through ATP syntase
proteins to make ATP from ADP P
127
Substrate level phosphorylation
- ATP is formed as a direct transfer of electrons
from the substrate to as ADP P? ATP
128
Oxidative phosphorylation
- Electrons made available in metabolism are
transferred to oxygen and ATP is produced in the
process. Chemiosmosis
129
(No Transcript)
130
(No Transcript)
131
(No Transcript)
132
(No Transcript)
133
(No Transcript)
134
(No Transcript)
135
Fermentation generates ATP by substrate level
phosphorylation. It is anaerobic Three
Types Alcoholic- Lactic Acid-
2NAD2Lactate
2 Ethanol 2CO2NAD
136
Photosynthesis
CO2H2O light CnH2n0nO2
Light- measured as an absorption spectrum, the
wavelengths that are most important are
different for different types of autotrophs
137
(No Transcript)
138
CO2
H2O
Light rxn.
Dark rxn.
light
Calvin Cycle
NADPH
PS1
Calvin Cycle
Thylakoid
ATP
PS2
Photolysis and Photophosphorylation
Stroma
O2
CnHnOn
139
(No Transcript)
140
Visible Spectrum
Reflected
Absorbed
Absorbed 680-700
141
(No Transcript)
142
Primary acceptor
NADP 2H
-e
pq
NADPH
Cytochrome complex
pc
Photosystem I P700
Chl a
700nm LIGHT
Photosystem II P680
680nm
143
Photosystem I
- Also known as P700-receives electrons from those
released in PSII to replace photoexcited
electrons uses light at far end of the red
wavelength - PSI 700
- PSII 680 the II in PSII H2O
144
Photosystem II
- P680
- due to an association with different proteins
- this system utilizes different wavelengths
- causes water to split capturing its electron
- it then transfers the electron to PSII
chlorophyll molecules
145
Noncyclic Electron Flow
Water is split (photolysis) and electrons pass
continuously from water to NADP. Primary
electron acceptors pass photoexcited electron to
the electron transport chain(Pq), (Pc),
cytochromes.
.
Uses both PSI and PSII
Generates O2, NADPH, and ATP
146
Cyclic Electron Flow
Excited Electrons pass through the electron
transport chain from P700 (PS I ) and return to
the starting point. Uses only PSI
Only ATP is generated
147
(No Transcript)
148
(No Transcript)
149
3CO2
Carbon Fixation
Calvin Cycle
RuBP
rubisco
6ATP
6ADP
3ATP
6NADPH
3ADP
6NADP
Regeneration RuBP
G3P--Glucose
150
Photorespiration
CO2 can act as a limiting factor. In cases where
there is not sufficient Carbon dioxide plants
will combine oxygen with RuBP to form
compounds that are broken down into CO2
151
Adaptations for Photosynthesis
- C4 Plants
- CO2 is added to PEP phosphophenolpyruvate
- stored in BUNDLE SHEATH CELLS near veins of leaf
- example- Corn
- CAM plants
- in hot dry areas plants must close stomates
- CO2 taken in at night is stored as an acid
152
Discovery of DNA
- Frederick Griffith
- Was studying Streptococcus Pneumonia
- Smooth vs. Rough Strains
- Smooth had a mucous coat and were pathogenic
(caused pneumonia) - Rough were non-pathogenic
- Conducted an experiment with mice
- Found out that the Rough bacteria became
transgenic
153
(No Transcript)
154
Discovery of DNA
- Avery, McCarty and MacLeod
- What was the genetic material in Griffiths
experiment? - Purified the heatkilled S-bacteria
- Into DNA, RNA, and Protein
- Mixed each with the R cells to see which one
transformed
155
Discovery of DNA
- Hershey-Chase Experiment
- Studied viruses that infect bacterial cells
- Bacteriophages
- Protein or DNA responsible for take-charge
actions of the virus? - Tagged the Protein with radioactive S
- Why?
- Tagged the DNA with radioactive P
- Why?
156
The Structure of DNAa double helix?
- Chargaffs Nucleic Acid Ratios
- Measured the base compositions of several species
- Percentage of each base present
- Human DNA
- A 30 and T 29
- G 20 and C 19
157
The Structure of DNAa double helix?
- Rosalind Franklin and Maurice Wilkins use X-Ray
diffraction to view structure - Watson and Crick propose a double helix using
their X-Ray pictures
158
DNA Double Helix
159
DNA Three Parts
- DNA is made up of nucleotides
- Nucleotides are made of
- Deoxyribose sugar
- Phosphate
- Base
- Guanine, Cytosine, Thymine and Adenine
160
DNA The Deoxyribose Sugar
161
DNA The Phosphate
162
DNA The Nitrogenous Bases
- Purines
- Adenine and Guanine
- Double Ring Structure
- Pyrimidines
- Thymine and Cytosine
- Single Ring Structure
163
Single Stranded DNA Nucleotides can only be
added to the 3 end of the nucleotide and
therefore addition of new nucleotides is always
5-----gt 3 DNA is anti-parallel!!
164
DNA STRUCTURE
165
How does it know to pair up?
- ADENINE ALWAYS PAIRS WITH THYMINE
- Two hydrogen bonds
- GUANINE ALWAYS PAIRS WITH CYTOSINE
- Three hydrogen bonds
166
Why do they pair up?
- Double helix had a uniform diameter
- Purine Purine
- too wide
- Pyrimidine Pyrimidine
- too narrow
- Purine Pyrimidine
- fits the x-ray data
167
One last look
Why does it twist?
168
DNA Replication
169
Meselson-Stahl demonstrate the Semiconservative
Replication of DNA using radioactive nitrogen
170
(No Transcript)
171
Why must DNA Replicate?
- Species Survival
- DNA must replicate BEFORE cell division
- Synthesis during Interphase
- All genes must be present in the daughter cells
172
Origins of Replication
- Sites along DNA that contain specific nucleotides
are recognized by specific proteins that initiate
process - In eukaryotes there are hundreds of thousands of
such points - Form replication bubbles
173
How does DNA Replicate?
- Hydrogen bonds break, forming bubbles
- Enzymes unwind and unzip
- Free nucleotides in the nucleus start process of
complementary base pairing - Nucleotides are fused together by DNA Polymerase
only 5 to 3 - Results in two identical double helixes
174
Replication Steps
- DNA helicase enzymes open double strand
- DNA uncoils and unzips exposing the DNA template
- Primase adds a RNA primer as binding proteins
hold strands together - DNA polymerase attaches to template at
replication fork - Nucleoside triphosphates add bases pairing A-T
and G-C as new strand is added to a 3 end,
primer removed
175
replication3
176
How does DNA Replicate?
177
How does DNA Replicate?
178
Leading Strand is Continuous
- A single RNA primers initiates the addition of
nucleotides to the 3 end of the leading strand
179
Lagging Strand
- Must wait for replication fork to open and then
add primer - Form Okazaki fragments
- RNA is removed only after addition of about 100
to 200 nucleotides - Fragments are joined by a ligase enzyme ( DNA
glue)
180
DNA Replication
181
The result
- DNA Replication results in TWO double helixes.
DNA unwinds and unzips, and new daughter strands
form, each complementary to an old parental
strand.
182
RNA - Structure
- Ribonucleic Acid different from DNA
- Always Single Stranded
- Ribose Sugar Base Unit
- Phosphate group (same in DNA)
- Nitrogenous Bases
- Cytosine always pairs with Guanine
- BUT! Adenine always pairs with URACIL
- (different in DNA!!!!!)
183
Four kinds of RNA
- Ribosomal RNA
- Messenger RNA
- Transfer RNA
- snRNA ribozyme in spliceosome
184
rRNA
- Ribosomal RNA or rRNA
- represents about 70 of cellular RNA
- joins with Ribosomal proteins to make the
cellular organelle RIBOSOMES - FUNCTION As a manufactured ribosome, supplies a
location for tRNA to join with mRNA to synthesize
a protein
185
mRNA
- 2. Messenger RNA or mRNA
- represents about 10 of cellular RNA
- contains the sequence of bases coding for a
particular amino acid sequence in a polypeptide
chain - removal of non-coding, internal sequences
(introns) - modification of the 5' base (cap)
- addition of adenines to 3' end (poly A tail)
- FUNCTION reads the DNA code (base sequence) and
becomes a copy that is read at the ribosome to
make a protein
186
hn RNA
- Pre-mRNA contains introns-non-coding regions as
well as exons-coding regions
187
Processing mRNA
- Deletion of introns
- Join exons
- Add cap ( GTP) and poly A tail
188
RNA splicing
- Spliceosome-several snurps
- snRNPs small nuclear
- Ribonucleoproteins-splicing enzyme( cuts and
glues)
189
Transcription
- DNA unzips at the locus of the gene being coded
- mRNA makes a copy of the gene
- Then
- mRNA is enzymatically modified
- A cap and a tail are added
- it then leaves the nucleus and finds a ribosome
(composed of rRNA and protein)
190
tRNA
- Transfer RNA or tRNA
- represents about 20 of cellular RNA
- each tRNA molecule is specific for one amino acid
- there is an enzyme for each amino acid which
recognizes the amino acid and its specific tRNA
and joins the two together - the specific joining of tRNA to amino acid is the
only place where the genetic code" applies - FUNCTION Pairs with Amino Acids and delivers
them to ribosomes at the right time to synthesize
a protein
191
Protein Synthesis
- Why should cells do this?
- Cells would not be able to grow and change
without proteins. - Proteins are found everywhere
- As enzymes, cell membranes, muscles, heart,
blood - What happens when proteins are not made correctly
or not made at all? - Ex. Cystic Fibrosis
- What part of DNA holds the code for the protein?
192
PROTEIN SYNTHESISEveryone is involved
- Transcription
- DNA, mRNA
- Translation
- mRNA, rRNA and tRNA
193
Pre-Translation
- mRNA binds to the ribosome
- Meanwhile, tRNAs are attaching to their amino
acids using tRNA Transferase - Free tRNAs, with their amino acids attached,
circulate in the cytoplasm and match up with the
triplet codes in the mRNA
194
Translation
- Initiation
- The first tRNA enters the ribosome at the A site
- The second tRNA enters the ribosome (at the P
site) and the amino acids are bonded together
PEPTIDE BOND - Elongation
- Both tRNAs shift in one direction and make room
for the next tRNA to enter the ribosome - this pattern continues until the protein is
complete - Termination
- Stop codon is read UAA, UAG, UGA
195
(No Transcript)
196
(No Transcript)
197
(No Transcript)
198
Quick Definitions
- A-site aminoacyl-tRNA binding site
- P-site peptidyl-tRNA binding site
- Triplet code DNA
- Codon RNA
- Anti-codon - tRNA
- Introns removed from initial mRNA
- Exons bonded together to make the finished mRNA
product for translation - Polyribosomes more than one ribosome reading
the mRNA at one time
199
Codons and anti-codons
- Triplet code on DNA TAC
- mRNA copies it CODON AUG
- tRNA carries the ANTICODON UAC
- The Genetic code reads the codon
- AUG, the amino acid Methionine
- 45 different anti-codons exist
- AUG is always the initiation codon
- GTP supplies energy needed to synthesize protein
- initiator tRNA always carries Methionine first!
- Initiation factors - proteins that bring all
parts together (mRNA, small subunit, large
subunit, and tRNAs)
200
Genetic Code
- Interprets what the DNA triplet code reads
- Is written in both DNA base language A, G, C, T
or RNA base language A, G, C, U - Determines the order for Amino Acids
- Is universal within all species
- Reads the same as the anti-codon (on tRNA) except
T is now U
201
Genetic Code
202
Gene Regulation
- Control of gene expression occurs at four levels
in human cells - Transcription and posttranscription control
(nucleus) - Translation and posttranslation control
(cytoplasm) - Various cells express different genes
- Genes can be turned on or off
- Genes respond to activity outside of the cell
- Control of transcription is most important
regulatory mechanism (binding factors and
enhancers) Presence of TF determines
specialization - Some binding factors are sensitive to hormones
203
DNA Technology
- Biotechnology or genetic engineering the use of
natural biological systems to produce a product
desired by human beings - Examples include
- Gene Cloning
- DNA Amplification
- Transgenic Organisms
- Gene Therapy
- Chromosome Mapping and Sequencing
204
Gene Cloning
205
Gene Cloning
- Recombinant DNA DNA from two different sources
(human and E. coli) - Plasmid circular DNA used to transport the gene
into the organism - Enzymes needed Restriction and Ligase
- Host cell usually bacteria, wall must become
competent in order for the bacteria to uptake the
plasmid - Restriction enzyme cleaves DNA and allows for DNA
fragment to insert at the sticky ends - Vector method of transporting a gene (virus,
plasmid)
206
pVIB lux genes
- 2 genes to produce LUCIFERASE
- Aldehyde (energy source) synthesis
- several genes
- Regulatory genes to turn of and on
207
DNA Amplification
- Polymerase Chain Reaction PCR
- Used to make multiple copies of the same gene
- Copies can be examined to see if they match any
other sources - Prevents constant extraction from the organism
and better results
208
(No Transcript)
209
(No Transcript)
210
(No Transcript)
211
(No Transcript)
212
Other Technologies
- Recombinant DNA - gene splicing
- Transgenic organism- an organism that contains
another organisms DNA
213
Transgenic Organisms
- Transgenic possessing gene(s) from another
organism - Gene Pharming Using transgenic farm animals to
produce pharmaceuticals - ex. CF, cancer, blood clots
- Genetically altering crops to be resistant to
insects and produce larger - http//biology.about.com/science/biology/gi/dynami
c/offsite.htm?sitehttp//abcnews.go.com/sections/
science/DailyNews/gmcorn5Fbutterflies000821.html - Suicide Genes
- Insulin
214
Gene Therapy
- Delivering the defective gene to the cells that
need it to produce a protein - Familial hypercholesteremia
- SCID severe combined immunodeficiency syndrome
(missing maturation enzyme for T and B cells)
215
Chromosome Mapping
- 100,000 human genes
- RFLPs Restriction Fragment Length Polymorphisms
used to probe a region of DNA visible under a
microscope - Restriction enzymes sequence AA
- Specific base digestion
- CF LAB
216
Human Genome Project
- HGP due for completion in 2002
- Already sequenced the Fruit Fly and E. Coli
217
(No Transcript)
218
Gene Therapy
- Delivering the defective gene to the cells that
need it to produce a protein - Cystic Fibrosis
- Vector method of transporting a gene (virus,
plasmid) - Mechanical - usually a laboratory tool used
(inoculating loop) - Biological - part or whole of an organism
(bacteria)
219
Chromosome Mapping
- 30,000 human genes
- RFLPs Restriction Fragment Length Polymorphisms
used to tag a region of DNA visible under a
microscope - Restriction enzymes sequence AA
- Specific base digestion
220
Sanger Method of DNA Sequencing
- 1. Heat DNA Strands until they separate
- 2. Add nucleotides and DNA Polymerase
- 3. Add Dedeoxynucleotides (A, T, G, and C) at
different time periods to stop replication - 4. Place fragments in to Gel Electrophoresis
- 5. Allow to migrate and read the Base Sequence
221
(No Transcript)
222
(No Transcript)
223
Human Genome Project
- HGP due for completion in 2002
- Already sequenced the Fruit Fly and E. Coli
- The ultimate goal of HGP is to associate human
traits and inherited diseases with particular
genes. - It promises to revolutionize both therapeutic and
preventive medicine techniques for many human
diseases.
224
Human Genome Project
- Genome - the complete collection of an organism's
genetic material. - The human genome is composed of an estimated
30,000 - A single human chromosome may contain more than
250 million DNA base pairs, and it is estimated
that the entire human genome consists of about 3
billion base pairs.
225
DNA Fingerprinting
- Treat suspects blood with the same restriction
enzyme - Place sample in Gel Electrophoresis
- Allow samples to migrate
- Compare the suspects with the blood found at the
crime scene - Used in Criminal Trials OJ Simpson
- OJ DNA was an exact match yet he was found not
guilty?