F1.Fo ATPase - PowerPoint PPT Presentation
1 / 19
Title:
F1.Fo ATPase
Description:
Suggests the H /ATP stoichiometry is non-integral. ... One-to-one stoichiometry. Exchange is accomplished by a single protein, the ADP/ATP carrier. ... – PowerPoint PPT presentation
Number of Views:825
Avg rating:3.0/5.0
Title: F1.Fo ATPase
1
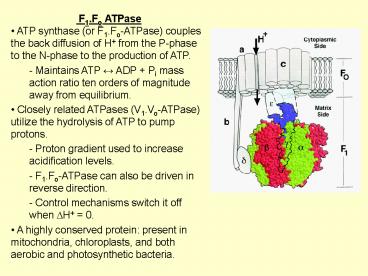
- F1.Fo ATPase
- ATP synthase (or F1.Fo-ATPase) couples the back
diffusion of H from the P-phase to the N-phase
to the production of ATP. - - Maintains ATP ? ADP Pi mass action ratio ten
orders of magnitude away from equilibrium. - Closely related ATPases (V1.Vo-ATPase) utilize
the hydrolysis of ATP to pump protons. - Proton gradient used to increase acidification
levels. - F1.Fo-ATPase can also be driven in reverse
direction. - Control mechanisms switch it off when DH 0.
- A highly conserved protein present in
mitochondria, chloroplasts, and both aerobic and
photosynthetic bacteria.
2
- Electron microscopy
- Observe knobs projecting from the matrix side of
the membrane. - When submitochondrial particles are washed with
urea the knobs were lost from the membrane. - The stripped submitochrondrial particles were
incapable of ATPsynthesis. - The membrane bound portion (Fo) acted as
proton-gradient uncoupler. - The knobs (or Fraction 1 F1-ATPase) could not
synthesize ATP but catalyzed the conversion of
ATP to ADP. - The active site for ATP synthesis lies in the
F1-ATPase domain. - - After reconstruction, ATP synthesis was
reproduced when incorporated with a proton pump. - ATP synthesis is coupled to proton pumping.
3
- Subunits of F1 ATP-synthase
- F1-ATPase (soluble) contains three copies of
subunit a and three of subunit b, and one each of
g, d and e. - Three active sites (one in each of the b
subunits) were recognized by sequence comparisons
and mutation studies. - Three additional ATP binding sites (one in each
of the a subunits) don't appear to be functional. - The b subunit may (sometimes) have ATPase
activity in the absence of the other subunits. - g subunit acts as a stalk with e associated.
- d subunit contributes to the stator.
4
- Subunits of Fo ATP-synthase
- Fo-ATPase contains one copy of subunit a, two
copies of subunit b, and ten (or eleven or twelve
....) copies of subunit c. - Contains one glutamic acid (or aspartic residue)
within an otherwise hydrophobic sequence - Believed to lie in the middle of the bilayer.
- Compare with other proton pumps, eg.
bacteriorhodopsin or cytochrome c oxidase. - All subunits are required to create a proton
pumping channel.
5
- Active site hydrolysis of ATP
- ATP hydrolysis is more easily studied than ATP
synthesis. - 18O labeled waters (oxygens in red) were used and
showed - At low ATP significant phosphate produced with
two 18 0 atoms.
6
- Boyer's interpretation
- The hydrolysis of ATP to ADP at the F1-ATPase
active site is (to some extent) reversible even
without input energy. - In solution the reverse reaction is
undetectably slow. - The rate of release of ADP from the active site
is slow relative to the rate of resynthesis of
ATP at the active site. - Tightly bound ADP and Pi form ATP with little
change in DG. - How can you make ATP without input energy?
- You don't make free ATP but rather bound ATP.
- ATP binding energy is perturbed so as to obey
thermodynamics (hence the 57 kJ/mol cost of ATP
production is recovered). - More complex experiments indicated ATP binding
at one site enhanced the release of ADP from
another. - - A conformational change in the protein changes
in the binding affinity of ATP so as to release
ATP.
7
- The structure of F1-ATPase
- The a and b subunits are arranged symmetrically
like an orange. - Subunit g passes through the middle.
- Three active sites observed with three different
substrates - ''Open'' empty site.
- ''Loose'' site, with bound ADP.
- ''Tight'' site, with bound AMP-PNP (a
non-hydrolysable ATP analogue). - Suggested that the enzyme operates by rotational
catalysis. - - Rotation of the g subunit inside the a3-b3
hexamer facilitates the binding of the substrate
and the release of the product.
8
- Observation of rotation of ATP
- Each b subunit was engineered to contain a large
his-tag, which was bound to a nickle surface. - The g subunit was engineered to contain a
biotinylated cysteine. - A flourescent actin rod (about 1 mu long) was
attached (through the biotin) to the subunit g. - In the presence of ATP the fluorescent rod
rotated (observed through a video
camera/microscope).
- - Without ATP present it moved randomly.
- Hydrolysis of ATP by F1-ATPase causes g subunit
to rotate. - - Fo-ATPase acts as a biological windmill.
- Rotation transferred by the g subunit and
enables ATP to be released from the active site.
9
Movie from a Japanese group
10
- Partial structure of Fo-ATPase
- Crystals structure from S. cerevisiae
mitochondria - An (almost) symmetric ring of 10 c subunits.
- Each subunit an a-helical hairpin (so have an
inner an outer ring of 10 a-helices) - The outer a-helices are slightly kinked inwards
at the centre.
11
- Interhelical loops of six to seven subunits in
close contact with the F1-ATPase g d central
stalk subunits. - - Suggests the c-ring stalk rotate together
during catalysis. - Subunits a b were not observed (lost during
crystallisation). - - No visible proton-translocation pathway.
- - No visible stator which counters the tendency
of a3-b3 to co-rotate with g.
12
- Side-chains could not be unambiguously assigned.
- - Length of helices cross-linking studies used.
- - Conserved Aspartate/Glutamate would lie about
halfway along the outer-ring a-helix. - Stock, Leslie Walker, Science 286, 1700-1705
(1999) (Most important references cited within
this paper).
13
- Mechanistic implications
- C-terminal a-helix contains the conserved
Asp/Glu essential for proton translocation. - - Probably lies on the outside surface the a/c
subunit interface forms the proton translocation
pathway. - Ten (rather than nine or twelve) c-subunits
visible in Fo-ATPase. - Suggests the H/ATP stoichiometry is
non-integral. - Non-intiger for F1 and Fo subunits suggests a
low-friction (no deep energy minima) rotation
mechanism. - Rotation fueled by the proton gradient.
- - Protonation/deprotonation pathways enabling the
c-ring to slip past the a-subunit could provide a
rotation mechanism.
14
- ATP/ADP Carrier
- ATP resynthesis occurs in the mitochondrial
matrix. - ATP is exported into the cytoplasm ADP is
imported into the matrix. - One-to-one stoichiometry.
- Exchange is accomplished by a single protein,
the ADP/ATP carrier. - Called mitochondria complex VI.
- Human bovine isoforms have 90 sequence
identity Yeast 50. - Functional dimer.
- All ADP/ATP carriers exhibit a consensus
sequence, RRRMMM, that is absent from other
mitochondrial carriers. - ADP/ATP carrier is a paradigm for mitochondrial
carriers.
.
15
- Structure of the ADP/ATP carrier
- Six transmembrane a-helices tilted relative to
the membrane to each other. - Form a barrel define a cone-shaped depression
accessible from the outside. - Cavity has a diameter of 20 Å and a depth of 30
Å. - The nucleotide carriers signature (RRRMMM) is
located at the bottom of this depression. - - Transport substrates bind to the bottom of the
cavity. - - Translocation requires a transition from a
pit to a channel conformation.
16
- Charge distribution
- Asymmetric distribution of charges within the
cavity. - ADP/ATP carrier signature, RRRMMM, spans the
thinnest part of the protein in a strategic
location for the transport. - Attraction of ADP towards the matrix against an
electrostatic potential is aided by the
distribution of positive charges within the
protein cavity.
17
- ADP ATP binding sites
- CATR (an inhibitor binds where ADP binds) was
located deep in the cavity off the
pseudo-threefold axis. - Bound by many hydrogen bonds.
- Tight binding of CATR explains why its a lethal
poison. - The conformation of the carrier for ATP binding
from the matrix is probably different from the
ADP-binding conformation. - - ATP binding site spectulated.
18
- Mechanism
- Functions as an active dimer.
- Each monomer can bind either ADP from the
outside or ATP or from the inside. - Transport takes place upon cooperative ADP ATP
binding. - Nucleotide binding favours binding of a second
nucleotide from the opposite side. - ATP binding can desabilize the salt bridges
induce conformational changes. - Prolines may act as hinges which straighten
odd-numbered helices pull open the channel. - The MMM motif occupies a bulky volume that may
act as a plug. - ADP/ATP transport in mitochondria also depends
upon the membrane potential nucleotide
concentrations.
19
- Unanswered questions
- What is the structure of the stator which
prevents the free rotation - of the F1 domain?
- What is the structure of the proton
translocation pathway? - How does this proton-pump act in reverse so as
to capture a proton wind'' and drive a
rotational motion? - Do underlying principles from bacteriorhodopsin/c
ytochrome c oxidase relate to proton pumping by
Fo-ATPase? - What is the second conformation of the ADP/ATP
carrier?