Discovering the structure of DNA - PowerPoint PPT Presentation
1 / 42
Title:
Discovering the structure of DNA
Description:
Mechanism of DNA repair ... A negative supertwists can be introduced that allow the break unwinding of the DNA ... The remaining DNA strand in turn serve as a ... – PowerPoint PPT presentation
Number of Views:131
Avg rating:3.0/5.0
Title: Discovering the structure of DNA
1
Discovering the structure of DNA
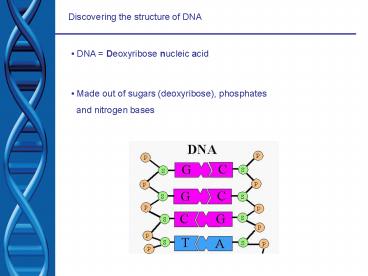
- DNA Deoxyribose nucleic acid
- Made out of sugars (deoxyribose), phosphates
- and nitrogen bases
2
Discovering the structure of DNA
- Structure was discovered in 1953 by James
- Watson and Francis Crick
3
Discovering the structure of DNA
Rosalind Franklins DNA image
Chargoffs rule A T C G
4
Cell division and DNA replication
- Cells divide
?Growth, Repair, Replacement
- Before cells divide they have to double cell
- structures, organelles and their genetic
- information
5
DNA replication
6
Site and function of nucleic acids
- DNA
- RNA
- site of DNA
- IN eukaryoytes cellsDNA is found in the
nucleus(chromosomes) and in the mitochondria. - In prokaryotes there is a single chromosome
which contain DNA .There may be also a non
chromosomal DNA in the form of plasmid. - Functions of DNA replications(cell division)
- expression of genetic information and protein
synthesis(through RNAs)
7
Site of RNAs
- 1. RNAs that synthesized in the mitochondria
remain within this organelle. - 2. RNAs that synthesized in the nucleus perform
their function in the cytoplasm. - Function of RNAs
- 1. RNAs participate in the process of expression
of genetic information that stored in DNA
(protein synthesis). - Some viruses use RNA in their its single or
double stranded form as a genetic material i.e
RNA is used instead of DNA.
8
Steps of DNA replication
- Replication folk
- chain elongation reverse transcriptase
- DNA repair
- regulation of DNA synthesis
- inhibitor of replication.
9
Semiconservative
- The process by which DNA is copied is called
semiconservative. This mean that after
replication ,each of daughter DNA mol. Will of
daughter DNA mol. Will contain - 1. one old strand one parental strand is
conserved. - 2.one new strand which is synthesized from free
nucleotide present in the nucleus.
10
Cont.
- During replication , the double strand DNA
mol.I(duplex) that is to be copied is separated
into two strands and each is used as a template
for the synthesis of a new complementary strand.
11
(No Transcript)
12
In prokaryotes
- DNA polymerase I catalyzes DNA repair.
- DNA polymerases II is unknown function.
- DNA polymerases III catalyses mostly replication
of DNA.
13
In eukaryotes
- DNA pol- alpha catalyses replication of nuclear
DNA. - DNA pol beta catalyses replication DNA repair.
- DNA pol gamma catalyses replication of
mtochondria DNA. - DNA pol delta responsible for leading strand of
DNA replication.
14
Cont.
- DNA pol e responsible for synthesis of lagging
strand and repair.
15
A. Strand separation
- For replication strands of DNA separated,
polymerase use only single stranded DNA as
template. - IN prokaryotes.E.coli ORIC initiation of
replication. - IN eukaryotes multiple site for replication
along the DNA helix.
16
Replication folk
- As strands unwind and separate , they form the
V shape where synthesis occur.This region is
called repliction folk. - 1. RF moves along the DNA mol. As synthesis
occur. - 2. replication of double stranded DNA is
bidirectional.
17
Proteins responsible
- A. helix destabilizing (HD) proteins they bind
nonenzymatically to a single stranded DNA,without
interfering with the ability of the nucleotides
serve as a template - Functions
- 1wo strands separated.
- Protect DNA from nuclease enzyme that cleave
single stranded DNA.
18
Cont.
- B. Helix unwinding proteins also called
helicase.or rep proteins. - 1. bind single stranded DNA near the replication
fork and then move into the neighbouring double
stranded region. - 2.Requires energy. 2ATP mol. Are consumed to
separate each base pair. - 3. once the strand separated destabilizing
proteins binds.
19
(No Transcript)
20
Topo I and II
- Swivels
- Prevents formation of supertwinsting and rotation
of the entire chromosome ahead of replication
folk. Super twisting makes further separation
more difficult and entire chromosome consume more
energy.
21
Topoisomerase I (DNA swivelases
- They cut and rejoin a single of double helix .
- This process does not require ATP as the energy
released from the cleavage (cutting ) of
phosphodiester bond is reused to reseal (rejoin)
the strand. - By creating transient nickthe DNA helix on
either side of the nick is allowed to rotate at
the phosphodiester bond opposite the nick ,thus
relieving accumulated supertwiste.
22
Topoisomerase II (DNA gyrase)
- It binds to both strand of the DNA and make
transient breaks in both strands of DNA helix to
pass through the break and finally reseal the
break . - A negative supertwists can be introduced that
allow the break unwinding of the DNA double helix.
23
Formation of RNA primer
- 1. polymerase III is unable to assemble the first
few nucleotides of a new strand using the parent
DNA strand as a template. - 2.This assembly require RNA primer
- A. a short fragment of RNA . 10 nucleotides.
- B.Complementary and antiparallel to the DNA
strand.
24
Cont.
- C. free -OH group at 3end . This Oh serves as a
the acceptor of the first nucleotide from DNA
polymerase III. - 3.synthesis of RNA primer requires primosome
which is a complex of an protein called RNA pol
and protein called DNA B protein. - Primosome binds with single stranded DNA and
enable the initiation of synthesis of RNA primer.
25
Cont.
- RNA primer is later removed.
26
Synthesis of new DNA
- The substrate of DNA are dATP,dGTP,dTTp,and
dCTP. If one of four nucleotide is not available
, DNA synthesis will blocked. - Using the free 3ÓH group of the RNA primer as the
acceptor of the first nucleotide , DNA polymerase
III begins to add subsequent nucleotide.
27
Chain elongation
- DNA pol lII moves along the template strand ,
substrate nucleotide pair with the pairing rule.
AT, GC,thus complementary to the parent strand. - New strand runs in 5-3 direction while template
strand runs 3-5. The daughter strand chain must
grow in opposite directions, one towards
replication fork and the other away from it.
28
Cont.
- Leading strand is the strand that being copied in
the direction towards replication fork . It is
synthesized almost continuously - Lagging strand is the strand being copied in
the direction away from replication fork . It is
synthesized discontinuously by forming small
fragment of DNA called OKAZAKI fragments. - They are joined to become a single , continuous
strand.
29
Excision of RNA primers and their replacement
with DNA
- 1. DNA polymerase III continues to synthesize DNA
un till it is blocked by a fragment of the RNA
primer. - 2.The RNA primer is excised by DNA polymerase I
- 3. Gaps resulting from the excised RNA primers
are filled by DNA pol I. - 4. Nicks are sealed by DNA ligase.
- 5. final phosphodiester linkage between the 5
phosphate group on the DNA chain synthesized by
DNA polymerase III and 3 hydroxyl group on the
chain made by DNA polymerase I is catalyzed by
DNA ligase .The energy required for this joint is
provided by cleavage of ATP tp AMP and Ppi.
30
Reverse transcriptase
- Also called RNA dependent DNA polymerase
- DNA RNA - protein
- Retroviruses has a mechanism for reversing the
first step in this flow form RNA to DNA. - The retrovirus contain ss RNA nucleic acid and a
viral enzyme called reverse transcriptase.
31
Mechanism of replication
- 1.Ss RNA ? ds DNA
- 2.This enzyme synthesize a DNA RNA hybride mol.
Using - A) RNA genome as template.
- B) dATP ,dGTP and dCTP gTTP as substrates.
- 3. RNA degraded by Rnase H .
- The remaining DNA strand in turn serve as a
template to form a double stranded genome of the
virus. - The newly synthesized viral double stranded DNA
enters the nucleus of the infected cell and can
integrate by recombination into host chromosome. - Eg HIV(AIDS) ,hepatitis A virus and some tumor
viruse. - RT are important in recombinant DNA technolongy.
32
DNA repair
- A)Causes of DNA damage
- Physical agent e.g x-ray , ultraviolet light.
- Chemical agent e.g alkylating agent
- Ionizing radiation
- B) single base alteration
- 1. depurination i.e removal of purine.
- 2.deamination of cytosine to uracil
- 3. deamination of adenine to hypoxathine.
- 4.Alkylation of base i.e addition of alkyl group.
- 5. Insertion or deletion of nucleotide.
- Base analog incorporation.
33
Two base alteration
- a. formation of thymine thymine dimer by
ultraviolet light
34
Cont.
- Chain break e.g phosphodiester bonds can be
broken. - Cross linkage
- A. between bases in same or opposite strands.
- B. between DNA and protein molecule e.g histones
35
fate of damaged DNA
- 1.Repaired
- 2. Replaced by DNA recombination
- 3. Retained retention leads to mutation and
cell death.
36
Mechanism of DNA repair
- Excision repair damaged only one strand eg
thymine dimers and missing base. - Repair of pyrimidine-pyrimidine dimer
- The dimers result form covalent joining of two
adjacent pyrimidine. - Caused by uv rays
- Thymine dimers prevent DNA pol from replicating
the DNA strand beyond the site of dimer
formation. - Dimer is excised and repair
- Uv specific endonuclease recognises the dimer
and makes a nick near it ,at 5 end - gap is filled by polymerase I ,in the direction
of 5 to 3.Other strand acts as template. - Thymine dimer region is excised by the 5-3
exonuclease activity of DNA pol I and sealed by
DNA ligase.
37
Xeroderma pigmentosum
- It is an autosomal recessive disease, is an e.g
of a defective mechanism for the repair of
pyrimidine dimers in DNA. - Absence of uv specific endonucleases require for
the recognition of the dimers is the cause of
this disease. - Individuals are sensitive to uv light which
causes extensive accumulation of thymine dimers
in skin cells with malignant transformation.
38
(No Transcript)
39
Some of the most common symptoms are An
unusually severe sunburn after a short sun
exposure. The sunburn may last for several weeks.
The sunburn usually occurs during a childs first
sun exposure. development of many freckles at an
early age. Irregular dark spots. Thin skin.
Excessive dryness. Rough-surfaced growths
(solar keratoses), and skin cancers. Eyes that
are painfully sensitive to the sun and may easily
become irritated, bloodshot, and clouded.
Blistering or freckling on minimum sun exposure.
Premature aging of skin, lips, eyes, mouth and
tongue
40
Repair of cytosine deamination to uracil
- Abnormal uracil is recognized by glycosylase that
cleaves the base . - Endonuclease cuts the phosphodiester bond on
5side. - DNA pol I fills the gap.
- DNA ligase seals the breaks.
41
Photoreactivation or light repair
- Thymine repair
- Use visible light (300-600nm) for activating
specific enzyme called photoactivating enzyme
which directly cleave and correct the dimer in
its place.
42
Recombination repair(sister strand exchange)
- In prokaryotes (E.coli) , the cell deal with DNA
replication at the dimer and reinitiating it on
the other side of the dimer . This leaves gap in
the newly synthesized strand b. - By sister strand exchange , the unmutated single
stranded segment from homologous DNA excised form
good strand (d strand) and inserted into the gap
present in b strand opposite the dimer. - The gap in d strand is next filled by polymerase
I.