Protein folding: Review of thermodynamics
1 / 16
Title:
Protein folding: Review of thermodynamics
Description:
Protein folding: Review of thermodynamics. Enthalpy : System Energy (Heat) ... Misfolding (by mutation) - nonfunctional proteins. Cystic fibrosis. p53 in cancer ... –
Number of Views:801
Avg rating:3.0/5.0
Title: Protein folding: Review of thermodynamics
1
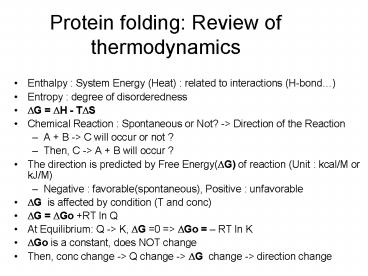
Protein folding Review of thermodynamics
- Enthalpy System Energy (Heat) related to
interactions (H-bond) - Entropy degree of disorderedness
- ?G ?H - T?S
- Chemical Reaction Spontaneous or Not? -gt
Direction of the Reaction - A B -gt C will occur or not ?
- Then, C -gt A B will occur ?
- The direction is predicted by Free Energy(?G) of
reaction (Unit kcal/M or kJ/M) - Negative favorable(spontaneous), Positive
unfavorable - ?G is affected by condition (T and conc)
- ?G ?Go RT ln Q
- At Equilibrium Q -gt K, ?G 0 gt ?Go RT ln K
- ?Go is a constant, does NOT change
- Then, conc change -gt Q change -gt ?G change -gt
direction change
2
Protein folding Review of thermodynamics
- Relationship between ?Go and K
- RT0.592 kcal/mol at 25degree
- ?Go RT ln K -0.592 ln K 1.36 log K
- Example A ? B with K10-6M ?Go -8.16
kcal/mol - A ? B with K 10-5M ?Go -6.8 kcal/mol
- Each 1.36 kcal/mol difference gives 10 fold
difference - in K and ration of reactant and product
Real example Hbond 6kcal/mol bonded vs
nonbonded van der Waals interaction
1kcal/mol Folded protein lt-gt unfolded protein
several kcal/mol (515) compare H-C covalent
bond 100 kcal/mol
3
Protein folding Review of thermodynamics
- Calculation of pKa with concentrations and pH
- Calculation of pH with concentrations and pKa
- Calculation of concentrations with pH and pKa
Example at pH 5.3, AcOH ACO- 110 at pH
6.3, AcOH ACO- 1100 Acetic acid solution -gt
pH adjust with NaOH to 6.3 -gt AcOH ACO- 1100
NaAc solid -gt pH adjust with HCl to 6.3 -gt AcOH
ACO- 1100 Equilibrium Does not matter where
you start NaH2PO4 Na2HPO4 -gt if we fix pH -gt
the equilibrium concentration is fixed (10mM
sodium phosphate buffer pH7.4)
4
Energetics of Protein folding
- ?G ?H - T?S
Unfolded state -gt folded state
To obtain negative dG, negative dH and positive
dS In reality H-bond (6kcal/mol), vdW
(1kcal/mol), ionic-interaction (1kcal/mol)
negative dH Folding Disordered state -gt ordered
state negative dS (conformational
entropy) Which effect is larger dH vs dS
5
Energetics of Protein folding
Secondary structure formation Large negative dH
(favorable) gt negative dS (unfavorable) regular
H bond formation favorable periodic ionic
interaction van der waals interaction alignment
of dipole Tertiary structure formation Not
much negative dH lt negative dS only some van der
Waals interaction or additional ionic
interaction Big decrease in protein
entropy Overall negative value of dG by
increase in Entropy of Water Hydrophobic
interaction entropy driven process water
molecules become ordered in direct contact with
hydrophobic molecule -gt decrease in entropy
Unfolded state -gt Folded
state (larger hydrophobic surface, large
H) (buried hydrophobic surface, small H) (less
disordered H2O, small S) (more disordered H2O,
large S) Therefore, protein folding increases S
(positive dS) of water molecules Overall, dG
values of protein folding process is quite small
(515kcal/mol) and mainly driven by water
entropy increase (hydrophobic interaction) Margina
l negative dG means that protein can be
dynamic Almost all the folded soluble proteins
have internal hydrophobic cores
?Swater (hydrophobic interaction)
- ?G
?G
?Sprotein
?Hprotein
5-15kcal/mol
6
Energetics of protein folding Hydrophobicity and
surface area
Hydrophobicity of an amino acid ?G aa in
octanol -gt aa in water ??G relative values to
Glycine
??G -3.07
W
W
Relationship between area and hydrophobicity Y
relative hydrophobicity (??G) X surface
area Blue residues Hydrophobic Red residues
Hydrophilic Blue residues the larger
hydrophobic surface , the larger ??G
(unfavorable) -gt hydrophobic surface corresponds
to free energy
22 cal/mol/Å2.
7
Energetics of Protein Folding
The native, folded structure of a protein, under
optimal conditions, is the most energetically
stable conformation possible
- Most of the information for determining the
three-dimensional structure of a protein is
carried in its amino acid sequence
Anfinsen, C.B. Principles that govern the folding
of protein chains. Science 181, 223-30 (1973).
8
Anfinsens Experiment
- Sequence determines structures
- Proof renaturation (ribonuclease by Anfinsen)
- Correct disulfide bonds
Transition state
dG
dG 5kcal/mol
Unfolded state
Folded state
Reaction process
9
Finding the most stable (Native) conformation
Folding Kinetics
- Protein synthesis unfolded state
- Final protein folded state (conformation)
- How to achieve folding?
- Folded state defined phi, psi angles
Lavinthals paradox Folding Speed
Kinetics E.coli (real world) 100 aa / 5
seconds Random folding (Assumption) Possible
conformation 10 phi, psi angles / residue -gt
10exp(100) times conformations 10exp(-13)
sec/conformation -gt 10exp(87)sec -gt 10exp(77)
years
Conclusion Protein folding is NOT a random
search (Not all the possibilities are
explored) There are specific pathways (shortcut)
-gt Kinteics
10
Models of shortcuts (folding pathway)
Hierarchical model Local secondary structure
folds first Long range interaction Hydrophobic
collapse Hydrophobic interaction -gt core
(non-local interaction) Molten globule
11
Protein folding funnel
Unfolded state Partially folded
state (Intermediates) Molten globule Folded
state
12
Protein Packing general rule
- Secondary structure pack closely, intercalation
- Polar or charged residues generally outside,
could be inside if charge is paired - Most hydrophobic residues inside
- Inside of protein 75 filled, 25 empty
- Loops, flexible parts outside, important
functional roles
13
Protein folding and stabilityFactors affecting
protein folding and stability
pH affect electrostatic interactions Temperature
Proteins are most stable (largest negative dG)
around 1530 degree Cold denaturation, Heat
denaturation, Thermophilic enzyme Detergent
Ionic, non-ionic (denaturating, non-denaturating)
Salts Hofmeister series changes water
entropy Cation NH4 gt K gt Na gt Li gt Mg2
gtCa2 gt (NH2)2CNH2 Anion SO42- gt HPO42- gt
CH3COO- gt citrate- gt tartarate- gt Cl- gt NO3- gt
I- gt SCN- Urea Disulfide bonds
14
Enzymes helping protein folding
- Chaperones
- Heat Shock Protein
- Binds hydrophobic regions
- Prevent aggregation
- Requires ATP hydrolysis
- Chaperonin
- GroEL/GroES
- protein disulfide isomerase,
- peptide prolyl isomerase
15
Folding enzymes and protein synthesis
16
Protein folding and diseases
- Misfolding (identical sequences) -gt insoluble
aggregates - Alzheimers disease
- Mad cow disease
- Creutzfeldt-Jakob
- Nobel Prize 1997, Stanley Prusiner
- (Prion Hypothesis)
- Protein is infectious
- Different folding (conformation) determines
infectivity - Misfolding (by mutation) -gt nonfunctional
proteins - Cystic fibrosis
- p53 in cancer
PrPc
?
PrPsc
PrPc? PrPsc (alpha) (high beta)
Trends in Biochemical Sciences Volume 29, Issue
4 , April 2004, Pages 162-165