CASE STUDY: Genetic Linkage Analysis via Bayesian Networks - PowerPoint PPT Presentation
Title:
CASE STUDY: Genetic Linkage Analysis via Bayesian Networks
Description:
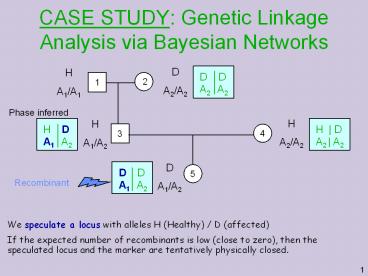
CASE STUDY: Genetic Linkage Analysis via Bayesian Networks D A2/A2 H A1/A1 2 1 Phase inferred H A2/A2 H A1/A2 3 4 D D A1 A2 H D A1 A2 H | D A2 | A2 D D – PowerPoint PPT presentation
Number of Views:123
Avg rating:3.0/5.0
Title: CASE STUDY: Genetic Linkage Analysis via Bayesian Networks
1
CASE STUDY Genetic Linkage Analysis via Bayesian
Networks
Phase inferred
We speculate a locus with alleles H (Healthy) / D
(affected) If the expected number of recombinants
is low (close to zero), then the speculated locus
and the marker are tentatively physically closed.
2
The Variables Involved
Lijm Maternal allele at locus i of person j.
The values of this variables are the possible
alleles li at locus i.
Lijf Paternal allele at locus i of person j.
The values of this variables are the possible
alleles li at locus i (Same as for Lijm) .
Xij Unordered allele pair at locus i of person
j. The values are pairs of ith-locus alleles
(li,li). The genotype Yj person I is
affected/not affected. The phenotype.
Sijm a binary variable 0,1 that determines
which maternal allele is received from the
mother. Similarly,
Sijf a binary variable 0,1 that determines
which paternal allele is received from the
father.
It remains to specify the joint distribution that
governs these variables. Bayesian networks turn
to be a perfect choice.
3
The Bayesian network for Linkage
This network depicts the qualitative relations
between the variables. We have already specified
the local conditional probability tables.
4
Details regarding recombination
L21m
L21f
L22f
L22m
S23m
X21
S23f
X22
Y2
Y1
L23f
L23m
X23
Y3
? is the recombination fraction between loci 2
1.
5
Details regarding the Loci
P(L11ma) is the frequency of allele a.
X11 is an unordered allele pair at locus 1 of
person 1 the data. P(x11 l11m, l11f) 0
or 1 depending on consistency
The phenotype variables Yj are 0 or 1 (e.g,
affected or not affected) are connected to the
Xij variables (only in the disease locus). For
example, model of perfect recessive disease
yields the penetrance probabilities
P(y11 sick X11 (a,a)) 1 P(y11 sick
X11 (A,a)) 0 P(y11 sick X11 (A,A)) 0
6
SUPERLINK
- Stage 1 each pedigree is translated into a
Bayesian network. - Stage 2 value elimination is performed on each
pedigree (i.e., some of the impossible values of
the variables of the network are eliminated). - Stage 3 an elimination order for the variables
is determined, according to some heuristic. - Stage 4 the likelihood of the pedigrees given
the ? values is calculated using variable
elimination according to the elimination order
determined in stage 3. - Allele recoding and special matrix multiplication
is used.
7
Comparing to the HMM model
S1
S2
S3
Si-1
Si
Si1
X1
X2
X3
Yi-1
Xi
Xi1
The compounded variable Si (Si,1,m,,Si,2n,f)
is called the inheritance vector. It has 22n
states where n is the number of persons that have
parents in the pedigree (non-founders). The
compounded variable Xi (Xi,1,m,,Xi,2n,f) is
the data regarding locus i. Similarly for the
disease locus we use Yi. REMARK The HMM
approach is equivalent to the Bayesian network
approach provided we sum variables
locus-after-locus say from left to right.
8
Experiment A (V1.0)
Elimination Order General Person-by-Person
Locus-by-Locus (HMM)
- Same topology (57 people, no loops)
- Increasing number of loci (each one with 4-5
alleles) - Run time is in seconds.
9
Experiment C (V1.0)
- Same topology (5 people, no loops)
- Increasing number of loci (each one with 3-6
alleles) - Run time is in seconds.
10
Some options for improving efficiency
- Multiplying special probability tables
efficiently. - Grouping alleles together and removing
inconsistent alleles. - Optimizing the elimination order of variables in
a Bayesian network. - Performing approximate calculations of the
likelihood.
11
Standard usage of linkage
There are usually 5-15 markers. 20-30 of the
persons in large pedigrees are genotyped (namely,
their xij is measured). For each genotyped person
about 90 of the loci are measured correctly.
Recombination fraction between every two loci is
known from previous studies (available genetic
maps). The user adds a locus called the
disease locus and places it between two markers
i and i1. The recombination fraction ? between
the disease locus and marker i and ? between the
disease locus and marker i1 are the unknown
parameters being estimated using the likelihood
function. This computation is done for every gap
between the given markers on the map. The MLE
hints on the whereabouts of a single gene causing
the disease (if a single one exists).
12
(No Transcript)
13
Parameter Estimation Lecture 10
Acknowledgement Some slides of this lecture are
due to Nir Friedman.
14
Likelihood function for a die Multinomial
sampling
Let X be a random variable with 6 values x1,,x6
denoting the six outcomes of a die. Suppose we
observe a sequence of independent outcomes
Data (x6,x1,x1,x3,x2,x2,x3,x4,x5,x2,x6)
What is the probability of this data ? If we
knew the long-run frequencies ?i for falling on
side xi, then,
15
Sufficient Statistics
- To compute the probability of data in the die
example we only require to record the number of
times Ni falling on side i (namely,N1, N2,,N6). - We do not need to recall the entire sequence of
outcomes - Ni i16 is called the sufficient statistics
for the multinomial sampling.
16
Sufficient Statistics
- A sufficient statistics is a function of the data
that summarizes the relevant information for the
likelihood - Formally, s(Data) is a sufficient statistics if
for any two datasets D and D - s(Data) s(Data ) ? P(Data?) P(Data?)
17
Maximum Likelihood Estimate
Maximum likelihood estimate is an assignment to
the parameters that maximizes the probability of
data (i.e., the likelihood function ). Usually
one maximizes the log-likelihood function which
is easier to do and gives an identical answer
18
Finding the Maximum
19
Adding Pseudo Counts
The MLE given by
can be misleading for small data sets because it
could happen that a small data set is not
typical. For example, it might be that we know
that the dice is manufactured to be loaded but
the small dataset we examined does not show this
property.
20
Example The ABO locus
Recall that a locus is a particular place on the
chromosome. Each locus state (called genotype)
consists of two alleles one parental and one
maternal. Some loci (plural of locus) determine
distinguished features. The ABO locus, for
example, determines blood type.
21
The ABO locus (Cont.)
However, testing individuals for their genotype
is a very expensive test. Can we estimate the
proportions of genotype using the common cheap
blood test with outcome being one of the four
blood types (A, B, AB, O) ?
The problem is that among individuals measured to
have blood type A, we dont know how many have
genotype a/a and how many have genotype a/o. So
what can we do ?
We use the Hardy-Weinberg equilibrium rule that
tells us that in equilibrium the frequencies of
the three alleles ?a,?b,?o in the population
determine the frequencies of the genotypes as
follows ?a/b 2?a ?b, ?a/o 2?a ?o, ?b/o 2?b
?o, ?a/a ?a2, ?b/b ?b2, ?o/o ?o2. So
now we have three parameters that we need to
estimate.
22
The Likelihood Function
Let X be a random variable with 6 values xa/a,
xa/o ,xb/b, xb/o, xa/b , xo/o denoting the six
genotypes. The parameters are ? ?a ,?b,
?o. The probability P(X xa/b ?) 2?a
?b. The probability P(X xo/o ?) ?o ?o.
And so on for the other four genotypes.
23
Computing MLE
- Finding MLE parameters nonlinear optimization
problem
P(Data ?)
?
24
Gene Counting
Had we known the counts na/a and na/o (blood type
A individuals), we could have estimated ?a from
n individuals as follows (and similarly estimate
?b and ?o)
Can we compute what na/a and na/o are expected to
be ?
We repeat these two steps until the parameters
converge.
25
Gene Counting (example of EM)
Input Counts of each blood type nA, nB, nO, nAB
of n people. Desired Output ML estimate of
allele frequencies ?a ,?b , ?o. Initialization
Set ?a ,?b ,and ?o to arbitrary values (say,
1/3). Repeat E-step (Expectation)