Using the genome - PowerPoint PPT Presentation
Title:
Using the genome
Description:
Using the genome Studying expression of all genes simultaneously Microarrays: reverse Northerns High-throughput sequencing Bisulfite sequencing to detect C ... – PowerPoint PPT presentation
Number of Views:138
Avg rating:3.0/5.0
Title: Using the genome
1
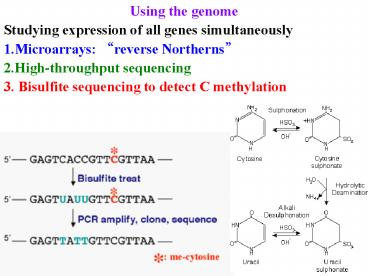
- Using the genome
- Studying expression of all genes simultaneously
- Microarrays reverse Northerns
- High-throughput sequencing
- Bisulfite sequencing to detect C methylation
2
Using the genome Bisulfite sequencing to detect C
methylation ChIP-chip or ChIP-seq to detect
chromatin modifications 17 mods are associated
with active genes in CD-4 T cells
3
- Generating the histone code
- Histone acetyltransferases add acetic acid
- Deacetylases reset by removing the acetate
4
- Generating the histone code
- CDK8 kinases histones to repress transcription
- Appears to interact with mediator to block
transcription - Phosphorylation of Histone H3 correlates with
activation of heat shock genes! - Phosphatases reset the genes
5
- Generating the histone code
- Rad6 proteins ubiquitinate histone H2B to repress
transcription - Polycomb proteins ubiquitinate histone H2A to
silence genes
6
- Generating the histone code
- Rad6 proteins ubiquitinate histone H2B to repress
transcription - Polycomb proteins ubiquitinate histone H2A to
silence genes - A TFTC/STAGA module mediates histone H2A and H2B
deubiquitination, coactivates nuclear receptors,
and counteracts heterochromatin silencing
7
- Generating the histone code
- Many proteins methylate histones highly
regulated!
8
- Generating the histone code
- Many proteins methylate histones highly
regulated! - Methylation status determines gene activity
9
- Generating the histone code
- Many proteins methylate histones highly
regulated! - Methylation status determines gene activity
- Mutants (eg Curly leaf) are unhappy!
10
- Generating the histone code
- Many proteins methylate histones highly
regulated! - Methylation status determines gene activity
- Mutants (eg Curly leaf) are unhappy!
- Chromodomain protein HP1 can tell the difference
between H3K9me2 (yellow) - H3K9me3 (red)
11
- Generating the histone code
- Chromodomain protein HP1 can tell the difference
between H3K9me2 (yellow) H3K9me3 (red) - Histone demethylases have been recently
discovered
12
- Generating methylated DNA
- Si RNA are key RNA Pol IV generates antisense or
foldback RNA, often from TE
13
Generating methylated DNA Si RNA are key RNA Pol
IV generates antisense or foldback RNA, often
from TE RDR2 makes it DS, 24 nt siRNA are
generated by DCL3
14
Generating methylated DNA RDR2 makes it DS, 24 nt
siRNA are generated by DCL3 AGO4 binds siRNA,
complex binds target Pol V
15
Generating methylated DNA RDR2 makes it DS, 24 nt
siRNA are generated by DCL3 AGO4 binds siRNA,
complex binds target Pol V Pol V makes
intergenic RNA, associates with AGO4-siRNA to
recruit silencing Complex to target site
16
Generating methylated DNA RDR2 makes it DS, 24 nt
siRNA are generated by DCL3 AGO4 binds siRNA,
complex binds target Pol V Pol V makes
intergenic RNA, associates with AGO4-siRNA to
recruit silencing Complex to target
site Amplifies signal! extends meth- ylated region
17
- Using the genome
- Many sites provide gene expression data online
- NIH Gene expression omnibus http//www.ncbi.nlm.ni
h.gov/geo/ provides access to many different
types of gene expression data
18
- Using the genome
- Many sites provide gene expression data online
- NIH Gene expression omnibus http//www.ncbi.nlm.ni
h.gov/geo/ provides access to many different
types of gene expression data - Many different sites provide digital Northerns
or other comparative analyses of gene expression - http//cgap.nci.nih.gov/SAGE
- http//www.weigelworld.org/research/projects/genee
xpressionatlas
19
- Using the genome
- Many sites provide gene expression data online
- NIH Gene expression omnibus http//www.ncbi.nlm.ni
h.gov/geo/ provides access to many different
types of gene expression data - Many different sites provide digital Northerns
or other comparative analyses of gene expression - http//cgap.nci.nih.gov/SAGE
- http//www.weigelworld.org/research/projects/genee
xpressionatlas - MPSS (massively-parallel signature sequencing)
http//mpss.udel.edu/
20
- Using the genome
- Many sites provide gene expression data online
- NIH Gene expression omnibus http//www.ncbi.nlm.ni
h.gov/geo/ provides access to many different
types of gene expression data - Many different sites provide digital Northerns
or other comparative analyses of gene expression - http//cgap.nci.nih.gov/SAGE
- http//www.weigelworld.org/research/projects/genee
xpressionatlas - MPSS (massively-parallel signature sequencing)
http//mpss.udel.edu/ - Use it to decide which tissues to extract our RNA
from
21
- Using the genome
- Many sites provide gene expression data online
- Many sites provide other kinds of genomic data
online - http//encodeproject.org/ENCODE/
22
- Post-transcriptional regulation
- Nearly ½ of human genome is transcribed, only 1
is coding - 98 of RNA made is non-coding
23
- Post-transcriptional regulation
- Nearly ½ of human genome is transcribed, only 1
is coding - 98 of RNA made is non-coding
- Fraction increases with organisms complexity
24
Known NcRNAs classes and functions
25
Implication in diseases
26
Implication in diseases
27
- Transcription in Eukaryotes
- 3 RNA polymerases
- all are multi-subunit
- complexes
- 5 in common
- 3 very similar
- variable unique ones
- Plants also have Pols IV V
- make siRNA
28
Transcription in Eukaryotes RNA polymerase I 13
subunits (5 3 5 unique) acts exclusively in
nucleolus to make 45S-rRNA precursor
29
- Transcription in Eukaryotes
- Pol I acts exclusively in nucleolus to make
45S-rRNA precursor - accounts for 50 of total RNA synthesis
30
- Transcription in Eukaryotes
- Pol I acts exclusively in nucleolus to make
45S-rRNA precursor - accounts for 50 of total RNA synthesis
- insensitive to ?-aminitin
31
- Transcription in Eukaryotes
- Pol I only makes 45S-rRNA precursor
- 50 of total RNA synthesis
- insensitive to ?-aminitin
- Mg2 cofactor
- Regulated _at_ initiation frequency
32
- Processing rRNA
- 100 bases are methylated
- C/D box snoRNA pick sites
- One for each!
33
- Processing rRNA
- 100 bases are methylated
- C/D box snoRNA pick sites
- One for each!
- 100 Us are changed to PseudoU
- H/ACA box snoRNA pick sites
- One for each!
34
- Processing rRNA
- 100 bases are methylated
- C/D box snoRNA pick sites
- 100 Us are changed to PseudoU
- H/ACA box snoRNA pick sites
- 3) Some snoRNA direct modification of tRNA and
snRNA
35
- Processing rRNA
- 200 bases are modified
- 2) 45S pre-rRNA is cut into 28S, 18S and 5.8S
products by ribozymes - RNase MRP cuts between 18S 5.8S
- U3, U8, U14, U22, snR10 and snR30 also guide
cleavage
36
- Processing rRNA
- 200 bases are methylated
- 2) 45S pre-rRNA is cut into 28S,
- 18S and 5.8S products
- 3) Ribosomes are assembled w/in nucleolus
37
RNA Polymerase III makes ribosomal 5S and tRNA
( some snRNA, scRNA, etc) gt100 different kinds
of ncRNA 10 of all RNA synthesis Cofactor
Mn2 cf Mg2 sensitive to high ?-aminitin
38
- Processing tRNA
- tRNA is trimmed
- 5 end by RNAse P
- (1 RNA, 10 proteins)
39
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- Some tRNAs are
- assembled from 2 transcripts
40
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- CCA is added to 3 end
- By tRNA nucleotidyl
- transferase (no template)
- tRNA CTP -gt tRNA-C PPitRNA-C CTP--gt tRNA-C-C
PPitRNA-C-C ATP -gt tRNA-C-C-A PPi
41
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- CCA is added to 3 end
- Many bases are modified
- Protects tRNA
- Tweaks protein synthesis
42
- Processing tRNA
- tRNA is trimmed
- Transcript is spliced
- CCA is added to 3 end
- Many bases are modified
- No cap! -gt 5 P
- (due to 5 RNAse P cut)
43
Splicing the spliceosome cycle 1) U1 snRNP
(RNA/protein complex) binds 5 splice site
44
SplicingThe spliceosome cycle 1) U1 snRNP binds
5 splice site 2) U2 snRNP binds
branchpoint -gt displaces A at branchpoint
45
SplicingThe spliceosome cycle 1) U1 snRNP binds
5 splice site 2) U2 snRNP binds
branchpoint -gt displaces A at branchpoint 3)
U4/U5/U6 complex binds intron displace
U1 spliceosome has now assembled
46
Splicing RNA is cut at 5 splice site cut end
is trans-esterified to branchpoint A
47
Splicing 5) RNA is cut at 3 splice site 6) 5
end of exon 2 is ligated to 3 end of exon 1 7)
everything disassembles -gt lariat intron is
degraded
48
SplicingThe spliceosome cycle
49
Splicing Some RNAs can self-splice! role of
snRNPs is to increase rate! Why splice?
50
Splicing Why splice? 1) Generate diversity
exons often encode protein domains
51
Splicing Why splice? 1) Generate
diversity exons often encode protein
domains Introns larger target for insertions,
recombination
52
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing
53
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing same gene encodes
different protein in different tissues
54
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing same gene encodes
different protein in different tissues Stressed
plants use AS to make variant stress-response
proteins
55
Why splice? 1) Generate diversity gt94 of human
genes show alternate splicing same gene encodes
different protein in different tissues Stressed
plants use AS to make variant Stress-response
proteins Splice-regulator proteins control AS
regulated by cell-specific expression and
phosphorylation
56
- Why splice?
- Generate diversity
- Trabzuni D, et al (2013)Nat Commun. 222771.
- Found 448 genes that were expressed differently
by gender in human brains (2.6 of all genes
expressed in the CNS). - All major brain regions showed some gender
variation, and 85 of these variations were due
to RNA splicing differences
57
- Why splice?
- Generate diversity
- Wilson LOW, Spriggs A, Taylor JM, Fahrer AM.
(2014). A novel splicing outcome reveals more
than 2000 new mammalian protein isoforms.
Bioinformatics 30 151-156 - Splicing created a frameshift, so was annotated
as nonsense-mediated decay - an alternate start codon rescued the protein,
which was expressed
58
Why splice? Splicing created a frameshift, so was
annotated as nonsense-mediated decay an
alternate start codon rescued the protein, which
was expressed Found 1849 human 733 mouse mRNA
that could encode alternate protein isoforms the
same way So far 64 have been validated by mass
spec
59
- Regulatory ncRNA
- SiRNA direct DNA-methylation via RNA-dependent
DNA-methyltansferase - In other cases direct RNA degradation
60
- mRNA degradation
- lifespan varies 100x
- Sometimes due to AU-rich 3'
- UTR sequences
- Defective mRNA may be targeted
- by NMD, NSD, NGD
- Other RNA are targeted by
- small interfering RNA
61
- Other mRNA are targeted by
- small interfering RNA
- defense against RNA viruses
- DICERs cut dsRNA into 21-28 bp
62
- Other mRNA are targeted by
- small interfering RNA
- defense against RNA viruses
- DICERs cut dsRNA into 21-28 bp
- helicase melts dsRNA
63
- Other mRNA are targeted by
- small interfering RNA
- defense against RNA viruses
- DICERs cut dsRNA into 21-28 bp
- helicase melts dsRNA
- - RNA binds RISC
64
- Other mRNA are targeted by
- small interfering RNA
- defense against RNA viruses
- DICERs cut dsRNA into 21-28 bp
- helicase melts dsRNA
- - RNA binds RISC
- complex binds target
65
- Other mRNA are targeted by
- small interfering RNA
- defense against RNA viruses
- DICERs cut dsRNA into 21-28 bp
- helicase melts dsRNA
- - RNA binds RISC
- complex binds target
- target is cut
66
- Small RNA regulation
- siRNA target RNA viruses ( transgenes)
- miRNA arrest translation of targets
- created by digestion of foldback
- Pol II RNA with mismatch loop
67
- Small RNA regulation
- siRNA target RNA viruses ( transgenes)
- miRNA arrest translation of targets
- created by digestion of foldback
- Pol II RNA with mismatch loop
- Mismatch is key difference
- generated by different Dicer
68
- Small RNA regulation
- siRNA target RNA viruses ( transgenes)
- miRNA arrest translation of targets
- created by digestion of foldback
- Pol II RNA with mismatch loop
- Mismatch is key difference
- generated by different Dicer
- Arrest translation in animals,
- target degradation in plants
69
- small interfering RNA mark specific
- targets
- once cut they are removed by
- endonuclease-mediated decay
70
(No Transcript)
71
- Most RNA degradation occurs in P bodies
- recently identified cytoplasmic sites where
exosomes XRN1 accumulate when cells are
stressed
72
- Most RNA degradation occurs in P bodies
- recently identified cytoplasmic sites where
exosomes XRN1 accumulate when cells are
stressed - Also where AGO miRNAs accumulate
73
- Most RNA degradation occurs in P bodies
- recently identified cytoplasmic sites where
exosomes XRN1 accumulate when cells are
stressed - Also where AGO miRNAs accumulate
- w/o miRNA P bodies dissolve!
74
- Thousands of antisense transcripts in plants
- Overlapping genes
75
- Thousands of antisense transcripts in plants
- Overlapping genes
- Non-coding RNAs
76
- Thousands of antisense transcripts in plants
- Overlapping genes
- Non-coding RNAs
- cDNA pairs
77
- Thousands of antisense transcripts in plants
- Overlapping genes
- Non-coding RNAs
- cDNA pairs
- MPSS
78
- Thousands of antisense transcripts in plants
- Overlapping genes
- Non-coding RNAs
- cDNA pairs
- MPSS
- TARs
79
- Thousands of antisense transcripts in plants
- Hypotheses
- Accident transcription unveils cryptic
promoters on opposite strand (Zilberman et al)
80
- Hypotheses
- 1. Accident transcription unveils cryptic
promoters on opposite strand (Zilberman et al) - 2. Functional
- siRNA
- miRNA
- Silencing
81
- Hypotheses
- 1. Accident transcription unveils cryptic
promoters on opposite strand (Zilberman et al) - 2. Functional
- siRNA
- miRNA
- Silencing
- Priming chromatin remodeling requires
transcription!