RATES OF NUCLEOTIDE SUBSTITUTION - PowerPoint PPT Presentation
1 / 19
Title:
RATES OF NUCLEOTIDE SUBSTITUTION
Description:
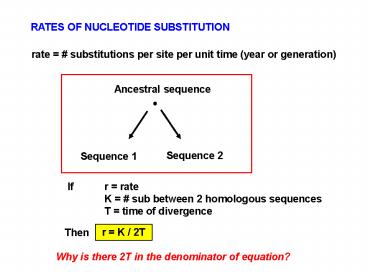
RATES OF NUCLEOTIDE SUBSTITUTION rate = # substitutions per site per unit time (year or generation) Ancestral sequence Sequence 2 Sequence 1 If r = rate – PowerPoint PPT presentation
Number of Views:101
Avg rating:3.0/5.0
Title: RATES OF NUCLEOTIDE SUBSTITUTION
1
RATES OF NUCLEOTIDE SUBSTITUTION
rate substitutions per site per unit time
(year or generation)
Ancestral sequence
Sequence 2
Sequence 1
If r rate K sub between 2 homologous
sequences T time of divergence
r K / 2T
Then
Why is there 2T in the denominator of equation?
2
In the literature, K or d (rather than r) is
often used for rate, (eg. Table 4.17, p.163)
so be sure to check context and units in figures
Within coding sequences, can separate the rates
into
KA amino acid-altering substitution rate
non-synonymous sub. / non-syn. site / year
KS synonymous substitution rate
synonymous sub. / syn. site / year
dN/dS term often used when comparing rates of
non-syn sub vs. syn sub
What are rates of nt substitutions between wheat
and maize nuclear genes (i) at synonymous sites
and (ii) at non-synonymous sites? p. 159-160
3
Rates of nt substitution in different parts of
genes
Fig. 4.3
4
1. Non-degenerate sites under greatest functional
constraint
- any change will alter protein
2. Sequences that are selectively neutral
evolve rapidly
eg pseudogenes, 4-fold degenerate, introns
- can be useful in molecular clock studies
3. Flanking (5) untranslated regions
somewhat constrained
- because contain gene regulatory signals
5
Similarity profile for 2 aligned DNA sequences
Fig. 4.4
6
Table legend Rates are in units of
substitutions per site per 109 years
Non-synonymous rates vary greatly among genes
(100-fold range), but synonymous rates are
relatively similar
KS usually much higher than KA
7
Ribosomal proteins - fundamental role in protein
synthesis - interact with rRNA and/or other rib.
proteins
Histones - fundamental role in DNA packing -
basic, compact proteins that interact with
DNA and/or other histones
Immunoglobulins - antibody diversity important
for immune response recognition of foreign
antigens
Fig. 4.6
For non-synonymous sites, the stronger the
functional constraint on aa sequence, the slower
the rate of evolution
8
Convert Table 4.1 information into Figure format
On log scale, plot values of KS and KA for S14
ribosomal protein insulin a-globin growth
hormone Ig k interferon b 1
10
102
103
104
105
Need to choose appropriate range for scale (and
units)
9
Non-synonymous substitutions rates within genes
Various functional (or structural) domains can be
subject to different constraints and evolve at
different rates
Fig. 4.5
10
Rate of nt substitution depends on
1. Functional constraints
- usually strong selective pressure against
changes that alter the protein, but cases of
positive selection
2. Mutational rate
- certain regions of genome may evolve at
different rates
Y chromosome sequences evolve 4 - 6 fold faster
than homologues on other chromosomes
Number of germ cell divisions for egg vs. sperm
production
males 200
females 33
DNA replication errors, lack of recombinational
repair
11
Positive selection (Adaptive evolution)
- rate of nonsynonymous substitution exceeds rate
of synonymous substitution
KA gt KS
Examples - some immunoglobulin genes (antibody
diversity)
- surface antigen genes of parasites and viruses
(evade host)
- some sex-related genes (for speciation,
reproductive barriers - to restrict gene flow?)
abalone sperm cell protein KA / KS 5.15 !!
But may be difficult to detect if - number of
substitutions is low - only one part of protein
under positive selection
12
Parallelism or molecular convergence
- independent occurrence of identical
substitutions - at homologous sites in different evolutionary
lineages - resulting in same phenotypic outcome
Example lysozymes in certain mammals (cow,
langur) and birds (hoatzin) adapted for activity
at low pH in posterior chamber of stomach
Fig. 4.8
13
Is ability of crocodile to stay underwater for
long times related to hemoglobin structure
evolution?
Comparison of crocodile (NC), alligator (MA),
caiman (SC) and human (HS) globin sequences
Komiyama et al. Nature 373244, 1995
14
By genetic engineering, changed several specific
codons in human globin gene, so amino acids
identical to crocodilian ones
- then measured O2 affinity of modified Hb
(a)
(b)
Oxygen-binding curves of crocodile Hb (circles)
and human Hb (squares) determined in the absence
(empty symbols) and presence (filled symbols) of
5 CO2.
(a) Wild-type Hb proteins (b) Human beta-chain
modified at positions 29, 31, 38, 39, 41
Komiyama et al. Nature 373244, 1995
15
These results indicate that an entirely new
function which enables species to adapt to a new
environment could evolve in a protein by a
relatively small number of amino acid
substitutions in key positions, rather than by
gradual accumulation of minor mutations.
Komiyama et al. Nature 373244, 1995
16
Possible reasons for variation in Ks among genes?
- if functional constraint at levels other than
amino acid sequence?
1. due to RNA folding? or cis-elements important
for RNA processing ?
2. codon usage bias?
If all codons specifying a particular amino acid
are functionally equivalent (selectively
neutral), expect similar frequencies
but observe non-random distribution, with
different patterns among organisms
17
Fig. 4.13
18
Correlation between codon usage pattern and
1. tRNA availability in cell (E. coli and yeast)
translational efficiency?
2. bias against certain dinucleotides (mutational
hotspots)
eg CpG in animal DNA
3. GC content of tightly-packed genomes
eg organellar, bacterial
19
Fig. 8.29