Genetics Maps - PowerPoint PPT Presentation
Title:
Genetics Maps
Description:
Title: Mapping the Human Genome Last modified by: Michael Scheid Created Date: 1/31/2005 3:24:50 PM Document presentation format: On-screen Show (4:3) – PowerPoint PPT presentation
Number of Views:197
Avg rating:3.0/5.0
Title: Genetics Maps
1
Genetics Maps
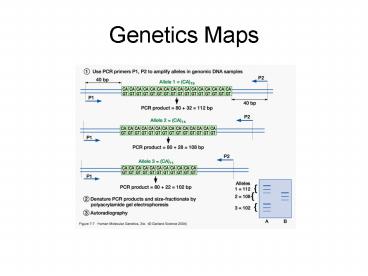
2
Genetics Maps
- Genotyping individuals with STRs
3
Genetics Maps
- By 1994, there was a 1 cM map based largely on
microsatellites (STRs)
A comprehensive human linkage map with
centimorgan density. Murray JC, Buetow KH, Weber
JL, Ludwigsen S, Scherpbier-Heddema T, Manion F,
Quillen J, Sheffield VC, Sunden S, Duyk GM, et
al.Science. 1994 Sep 30265(5181)2049-54.
5840 loci total 3617 polymerase chain
reaction-formatted short tandem repeat
polymorphisms 427 genes 0.7 centimorgan density
4
Genetics Maps
5
Genetics Maps
6
Physical Maps
7
Physical Maps
Ordering clones based on Hybridization
8
Physical Maps
Ordering clones based STS content
9
Genetic Maps and Physical Maps are Aligned by STS
10
Sequencing the Human Genome
ddG
ddT
ddA
ddC
3
Gel run
5
11
Sequencing the Human Genome
12
Sequencing the Human Genome
13
Sequencing the Human Genome
- Sequence 500 bp each reaction
- To sequence the Human genome, sequencing method
needs to be - FAST
- CHEAP
- In 1990 reality was
- SLOW
- EXPENSIVE (gt1 per base!)
14
Sequencing the Human Genome
- International Human Genome Sequencing Consortium
- Primarily six institutes with high-throughput
sequencing capabilities - Whitehead Institute
- The Sanger Center
- Washington University
- DOE Sequencing Center
- Bayer College of Medicine (31 Jan 2005 161,489
kb 2,935,479 kb) - In 1990, the IHGSC began a 15 year plan to
sequence the entire Human genome
15
Sequencing the Human Genome
16
Sequencing the Human Genome
- IHGSC Strategy - Shotgun sequencing of ordered
BAC contigs - Define BAC contig order based on STS
- Sequence each cluster of BACs within contig
align based on sequence - Anchor to genome by STS.
17
Sequencing the Human Genome
18
Sequencing the Human Genome
- In 1998, Celera Genomics announced plans to
sequence the human genome - 175,000 sequence reads per day, operating 24
hours a day, 7 days a week
J. Craig Venter
19
Sequencing the Human Genome
- Whole genome shotgun approach vs. Clone by Clone
approach - By-passes the initial work of ordering clones
- Celera performed about 32 million sequence reads,
each 500 1000 bp
20
Sequencing the Human Genome
21
Sequencing the Human Genome
- IHGSC published sequence reads every 24 hours to
prevent patenting of DNA - Celera had access to IHGSC data
- Debate over whether Celera could have shotgun
sequenced the genome without IHGSC data
22
Sequencing the Human Genome
- Both groups published results simultaneously
- Celera Science
- February 2001
- IHGSC Nature
- February 2001
23
Sequencing the Human Genome
Nature 409, 818 - 820 (15 February 2001)
24
Sequencing the Human Genome
- Controversy! Science published Celeras sequence
without requiring deposition to GenBank - Celera provides full access, with a catch
- Celera provided Science with a copy in escrow
25
(No Transcript)
26
Sequencing Your Human Genome
- For 500,000 you can have your DNA sequenced
- Sequence 1000 individual human genomes
- Personalized medicine
J. Craig Venter
27
- Next Gen Sequencing
28
- The proliferation of genetic testing resources
29
Human Genome
- Legal considerations
- Should DNA, or genes, be patentable?
- In the past, USPTO considered genes as man-made
chemicals - Copy DNA region, splice it together, and
propagate it in bacteria, etc
30
Human Genome
- Celera gt6500 genes
- Human Genome Sciences gt7000
- Incyte gt50,000
- Only a fraction may be awarded by USPTO, and only
a fraction of these may be useful in treating
human disease
31
Human Genome
- 1994 U. of Rochester scientists isolate mRNA for
COX-2 and clone gene - Suggest that compounds which inhibit COX-2 might
provide pain relief from arthritis - Submit patent application in 1995
32
Human Genome
- 1998 Celebrex inhibitor of cyclooxygenase-2
(COX-2) introduced as arthritis medication - Developed by Pfizer/Searle
- Development began in early-90s i.e. around time
of U. of Rochester discovery
33
Human Genome
- April 2000, U. of Rochester awarded patent
covering COX-2 gene and inhibition of the peptide
product thereof - The same day, U. of Rochester files lawsuit
against Pfizer/Searle to block Celebrex sales - Claims that Pfizer/Searle infringes on their
patent - They want royalties from the sale of the invention
34
Human Genome
- 2003 U. of Rochester patent found invalid
- 2004 Invalidation upheld by higher Court
- U. of Rochester patent did not provide sufficient
example of what the inhibitor would bei.e.
claims too broad without a working example - How will basic science performed by
Universities be rewarded?
35
Human Genome
- Vioxx and Celebrex in news again this year
increased risk of cardiovascular event i.e.
heart attacks
36
Human Genome
- Gene discovery
- Average gene extends over 27 kb
- Average 8.8 introns
- Average 145 bp
- Extremes
- Dystrophin gene 2.4 Mb
- Titin gene contains 178 introns, coding for a
80,780 bp mRNA
37
Human Genome
- Gene discovery
- One approach is to examine transcriptome
- Exome
38
Human Genome
- Conservation of chromosome/gene location between
organisms - Synteny
- Exons tend to be conserved between species
39
Human Genome
- Human vs. Pufferfish genome
- Pufferfish genome about 1/7th the size of the
human genome with similar number of genes
40
Human Genome
- Predictive computer programs, e.g. GENSCAN
- GENSCAN predicts the location of genes based on
splicing predictions, promoter regions and other
criteria
41
Human Genome
- Online databases have formed to curate Human
genome data - Ensembl (www.ensemble.org)
42
Genetic Mapping of Mendelian Characters
43
Identifying Disease-Causing Gene Variations
- Linkage analysis and Positional Cloning
- Clone disease gene without knowing anything
except the approximate chromosomal location
44
Recombination
- Recombination during meiosis separates loci
- More often when they are farther apart
- Less often when they are close
- Recall discussion of the Genetic Map
- Loci on separate chromosomes segregate
independently - Loci on the same chromosome segregate as a
function of recombination
45
Recombination
13-1
46
13_06.jpg
47
Linkage analysis
- Linkage analysis locates the disease gene locus
- Linkage analysis requires
- Clear segregation patterns in families
- Informative markers close to the locus
- Utilize LOD analysis to verify linkage
- Calculate cM distance between Loci
48
Positional Cloning
- Widely used strategy in human genetics for
cloning disease genes - No knowledge of the function of the gene product
is necessary - Strong for finding single-gene disorders
49
Positional Cloning
- Linkage analysis with polymorphic markers
establishes location of disease gene - LOD score analysis, and other methods are
employed - Once we know the approximate location
- The heavy molecular biology begins
50
Positional Cloning
- Example - Huntingtons disease
- CAG
- Autosomal dominant
- 100 penetrance
- Fatal
- Late onset means patients often have children
51
Finding the Huntington Gene 1981-1983
- Family with Huntington's disease found in
Venezuela - Originated from a single founder - female
- Provided
- Traceable family pedigree
- Informative meiosis
- Problem was only a few polymorphic markers where
known at the time
52
Finding the Huntington Gene
- Blood samples taken
- Check for disease symptoms
- Paternity verified
53
Finding the Huntington Gene
- By luck, one haplotype segregated very closely
with Huntington disease - Marker was an RFLP called G8 (later called D4S10)
54
(No Transcript)
55
Finding the Huntington Gene
56
Finding the Huntington Gene
- Locate the region to the tip of the short arm of
chromosome 4 by linkage with G8 (D4S10) - Maximum LOD score occurred at about 4 cM
distance, i.e. 4 in 100 meiosis
57
Finding the Huntington Gene
- Together this started an international effort to
generate YAC clones of the 4 Mb region - More polymorphisms were found
58
Finding the Huntington Gene
- Next, find an unknown gene in an uncharacterized
chromosome location - Locate CpG islands
- Cross-species comparisons
- Further haplotype analysis suggested a 500 Kb
region 3 to D4S10
59
Finding the Huntington Gene
- Exon trapping was key
- Compare cloned exons between normal and
Huntington disease patients
60
Finding the Huntington Gene
61
Finding the Huntington Gene
- One exon, called IT15, contained an expanded CAG
repeat. - Mapping to 4 cM 1983
- Cloning of Huntington gene 1993
62
Complex Disease and Susceptibility
Single gene disorders Mendelian
Inheritance High penetrance Low environmental
influence (but sometimes significant) LOD-based
linkage analysis works great Genetic
heterogeneity Low population incidence
Gene
Gene
Disease
63
Complex Disease and Susceptibility
Gene
Gene
Gene
Gene
Environment
Disease A
Disease B
Disease C
Multifactorial disorders