DICOM Network Roles - PowerPoint PPT Presentation
1 / 25
Title:
DICOM Network Roles
Description:
Title: Seminario Geant4 INFN Author: bc+sc Last modified by: BC Created Date: 5/8/1997 12:59:37 AM Document presentation format: On-screen Show Other titles – PowerPoint PPT presentation
Number of Views:108
Avg rating:3.0/5.0
Title: DICOM Network Roles
1
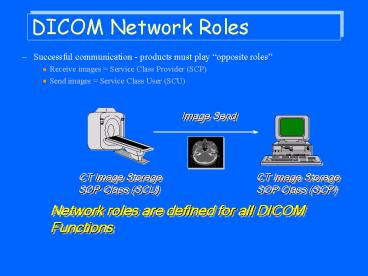
DICOM Network Roles
- Successful communication - products must play
opposite roles - Receive images Service Class Provider (SCP)
- Send images Service Class User (SCU)
CT Image StorageSOP Class (SCU)
Network roles are defined for all DICOM Functions
2
DICOM Conformance Statement
- It is Required!
- It is a Public Document
- It Conveys a Products DICOM Functionality
- It is Based on DICOM Vocabulary
- Abstract Syntaxes (SOP Classes), Transfer
Syntaxes, SCU/SCP.. - It is Used to Compare Connectivity
- It is most Often on the Web _at_ Vendor Site
- It Does Not Address All of an Applications
Capabilities, but should Address All of the
Applications DICOM ones
A Major Step Towards Interoperability
3
Language and dictionary
128 bit inutili (non sempre) DICM
4
std DICOM
5
DICOM
- (0008,0000) UL 424 4 IdentifyingGroupLength
- (0008,0020) DA 20070509 8 StudyDate
- (0008,0022) DA 20070509 8 AcquisitionDate
- (0008,0023) DA 20070509 8 ImageDate
- (0008,0030) TM 163853 6 StudyTime
- (0008,0032) TM 163933 6 AcquisitionTime
- (0008,0060) CS CT 2 Modality
- (0008,0070) LO Philips 8 Manufacturer
- (0008,0080) LO OSP S.CROCE CUNEO 18
InstitutionName - (0008,1040) LO RADIOLOGIA 10
InstitutionalDepartmentName - (0008,1090) LO Brilliance 64 14
ManufacturerModelName - (0010,0010) PN TEST CTDI 16 CM HEAD. 22
PatientName - (0010,0020) LO 11111 6 PatientID
- (0010,0030) DA 20070509 8 PatientBirthDate
- (0010,0040) CS O 2 PatientSex
- (0018,0050) DS 0.5 6 SliceThickness
- (0018,0060) DS 120 4 KVP
- (0018,0090) DS 500 4 DataCollectionDiamete
r - (0018,1030) LO Encefalo Ax Testa 22
ProtocolName
6
Images dimension
- Ammettendo che ogni voxel occupi 32 byte qual è
lo spazio RAM necessario a simulare 40 cm di TAC
di cui il seguente è parte dellheader DICOM di
unimmagine? - circa 170 MByte
- circa 2 Mbyte
- circa 670 Mbyte
7
Geant4-DICOM interface
- Developed by L. Archambault, L. Beaulieu, V.-H.
Tremblay (Univ. Laval and l'Hôtel-Dieu, Québec) - Donated to Geant4 for the common profit of the
scientific community - under the condition that further improvements and
developments are made publicly available to the
community - Released with Geant4 5.2, June 2003 in an
extended example - by S. Guatelli mainly
- Deeply revised in by Pedro Arce in 2007
- Small improvements by S. Chauvie
T. Aso A.Kimura Ashikaga Institute of
Technology
Geant4 examples/extended/medical/DICOM
8
From phantom to MC
9
cont
-
Density
Range Materials
-------------------------------------------------
-- g/cm3
- ----------------------
-----------------------------
0.100 , 0.351 Lungs (inhale)
0.351 , 0.800
Lungs (exhale) 0.919 , 0.979
Adipose
0.979 , 1.004 Breast
1.004 , 1.043
Phantom 1.043 , 1.109
Liver
1.109 , 1.113 Muscle
1.113 , 1.400
Trabecular Bone 1.496 , 1.654
Dense Bone
ICRU 46
10
and ends.
11
The structure
12
From DICOM image to Geant4 geometry
- Reading image information
- Transformation of pixel data into densities
- Association of densities to a list of
corresponding materials - Defining the voxels
- Geant4 parameterised volumes
- parameterisation function material
13
Start reading DICOM files
14
Tranlsate TAGS with DICT
15
Read the header and create the tag
16
Implicit Endian Explicit VR, special cases
17
Implicit Endian Explicit VR, other cases
18
Implicit Endian Implicit VR
19
Create .g4dcm
20
Data.dat
21
CT2Density.dat
22
Write density
Splitting materials in density intervals In the
class DicomDetectorConstruction, it is defined a
density interval G4double densityDiff 0.1
23
Navigation.
- The 1D optimisation . It will be very slow
because each time a track exits a voxel it has to
loop to all other voxels to know which one it may
enter - The 3D optimisation with G4SmartVoxel a 3D grid
is built, so that the location of voxels is fast,
but it requires a lot of memory - Using G4NestedParameterisation. The search is
done hierarchically in X, Y and Z. It is fast and
does not require big memory - Using G4PhantomParameterisation/G4RegularNavigati
on an special algorithm to navigate in regular
voxelised geometries (see GEANT4 doc). This is
the fastest way without any extra memory
requirement (and it is the default in this
example). It includes an option (default) to skip
frontiers between voxels when they have the same
material. When using this option at each step the
energy is all deposited in the last voxel for
properly distribution of the dose
(energy/volume) the G4PSDoseDeposit_RegNav
scorer can be used
24
MC Dose calculation in Radiotherapy
How accurate is the dose calculation ?
Description of patients
Energy deposition
Physics
Geometry
Validation studies
DICOM
25
Grazie per lattenzione!