Next step: deciding which genes to clone - PowerPoint PPT Presentation
Title:
Next step: deciding which genes to clone
Description:
Topic Next step: deciding which genes to clone Problem = correlating enzymes with genes Who matches the pH? Who localizes where? Which isoform if alternatively spliced? – PowerPoint PPT presentation
Number of Views:47
Avg rating:3.0/5.0
Title: Next step: deciding which genes to clone
1
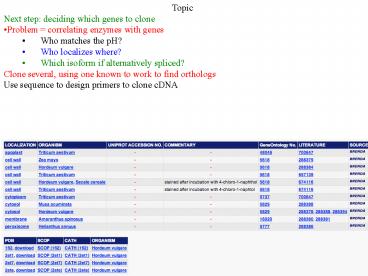
- Topic
- Next step deciding which genes to clone
- Problem correlating enzymes with genes
- Who matches the pH?
- Who localizes where?
- Which isoform if alternatively spliced?
- Clone several, using one known to work to find
orthologs - Use sequence to design primers to clone cDNA
2
- Issues
- All genes that have been cloned and expressed
have acidic pH optima but Trinchant found
bacteroid with pH 8 opt - Oxalate oxidase belongs to the cupin superfamily
- functionally diverse proteins that have a highly
conserved pattern of two histidine-containing
motifs the cupin signature
3
Cupins functionally diverse proteins that have a
highly conserved pattern of two
histidine-containing motifs the cupin
signature Proposal you each clone one safe and
one risky oxalate ox Safe known Oxalate oxidase
or decarboxylase Risky based on homology
4
- Cupins
- functionally diverse proteins that have a highly
conserved pattern of two histidine-containing
motifs the cupin signature - Proposal you each clone one safe and one risky
oxalate ox - Safe known Oxalate oxidase or decarboxylase
- Wheat oxalate oxidase GF-2.8 P15290http//www.ncb
i.nlm.nih.gov/pmc/articles/PMC1987392/ - Barley oxalate oxidase CAA74595
- Rice oxalate oxidase Os03g0693700 or
Os03g0694000 - Ceriporiopsis subvermispora oxalate oxidase
AJ746414 - B. subtilis oxalate decarboxylase O34714
- White rot oxalate decarboxylase AF200683
5
Risky based on homology, eg to rice
OXO1 SORBIDRAFT_01g011370 Sorghum
bicolor Sequence ID refXP_002464052.1
Length 225 Expect Identities
Positives Gaps 1e-123
171/216(79) 196/216(90) 0/216(0) LOC100272932
precursor Zea mays Sequence ID
refNP_001140856.1 Length 225 Expect
Identities Positives
Gaps 3e-122 173/228(76) 203/228(89)
4/228(1) oxalate oxidase GF-2.8-like
Brachypodium distachyon Sequence
ID refXP_003561035.1 Length 226 Expect
Identities Positives
Gaps 4e-114 168/222(76) 191/222(86)
4/222(1)
6
Risky based on homology, eg to Bacillus
subtilis oxdC as query Accession
NP_391204.1oxalate decarboxylase oxdC
Neurospora crassa OR74ASequence ID
refXP_964781.1Length 455Expect Identities
Positives Gaps5e-150
211/356(59) 269/356(75) 2/356(0)Oxalate
decarboxylase Mesorhizobium sp.
LNHC209A00Sequence ID refWP_023795524.1Length
377Expect Identities Positives
Gaps8e-150 210/367(57)
259/367(70) 4/367(1)hypothetical protein
BJ6T_62210 Bradyrhizobium japonicumSequence
ID refYP_005611058.1Length 357Expect
Identities Positives
Gaps 3e-180 244/355(69) 283/355(79)
1/355(0)
7
Risky based on homology, eg to Agrobacterium
oxalate decarboxylase Accession NP_355894.1OxdD
Rhizobium sp. IRBG74Sequence ID
refYP_008649166.1Length 415Expect Identities
Positives Gaps0.0
402/415(97) 406/415(97) 0/415(0)cupin
Rhizobium lupiniSequence ID
refWP_006700522.1Length 415Expect Identities
Positives Gaps0.0
404/415(97) 407/415(98) 0/415(0)Oxalate
decarboxylase Bradyrhizobium sp. Sequence ID
gbEJZ30017.1Length 406Expect Identities
Positives Gaps0.0
267/403(66) 320/403(79)
2/403(0)oxalate decarboxylase Bradyrhizobium
japonicum USDA 6Sequence ID refYP_005611126.1
Length 415Expect Identities
Positives Gaps0.0
264/413(64) 321/413(77) 3/413(0)
8
Risky based on homology, eg to Agrobacterium
oxalate decarboxylase Accession
NP_355894.1 uncharacterized protein LOC102383685
Alligator sinensis XP_006018933.1
XM_006018871 Length 341 Expect 5e-12,
Identities 67/277 (24), Positives 119/277
(42), Gaps 28/277 (10) uncharacterized
protein C18orf54 Bos taurus XP_005224336.1
XM_005224279 Length 453 Expect
7e-12,Identities 78/312 (25), Positives
132/312 (42), Gaps 36/312 (11)
9
How to proceed?
Kinetic and Spectroscopic Studies of Bicupin
Oxalate Oxidase and Putative Active Site Mutants
10
Using the genome Bisulfite sequencing to detect C
methylation ChIP-chip or ChIP-seq to detect
chromatin modifications 17 mods are associated
with active genes in CD-4 T cells
11
- Using the Genome
- Acetylation, egH3K9Ac, is associated with active
genes - Phosphorylation shows condensation
- Ubiquitination of H2A and H2B shows repression
- Methylation is more complex
- H3K36me3 on
- H3K27me3 off
- H3K4me1 off
- H3K4me2 primed
- H3K4me3 on
12
- Histone code
- Modifications tend to group together genes with
H3K4me3 also have H3K9ac - Cytosine methylation is also associated with
repressed genes
13
- Generating the histone code
- Histone acetyltransferases add acetic acid
14
- Generating the histone code
- Histone acetyltransferases add acetic acid
- Many HAT proteins mutants are very sick!
15
- Generating the histone code
- Histone acetyltransferases add acetic acid
- Many HAT proteins mutants are very sick!
- HATs are part of many complexes
16
- Generating the histone code
- Bromodomains specifically bind acetylated lysines
17
- Generating the histone code
- Bromodomains specifically bind acetylated lysines
- Found in transcriptional activators general TFs
18
- Generating the histone code
- acetylated lysines
- Deacetylases reset by removing the acetate
19
- Generating the histone code
- acetylated lysines
- Deacetylases reset by removing the acetate
- Deacetylase mutants are sick!
20
- Generating the histone code
- Deacetylases reset by removing the acetate
- Deacetylase mutants are sick!
- Many drugs are histone deacetylase inhibitors
21
Generating the histone code Deacetylases reset
by removing the acetate Deacetylase mutants are
sick! Many drugs are histone deacetylase
inhibitors SAHA suberanilohydroxamic acid
vorinostat Merck calls it Zolinza, treats
cutaneous T cell lymphoma
22
Generating the histone code Deacetylases reset
by removing the acetate Deacetylase mutants are
sick! Many drugs are histone deacetylase
inhibitors SAHA suberanilohydroxamic acid
vorinostat Merck calls it Zolinza, treats
cutaneous T cell lymphoma Binds HDAC active site
chelates Zn2
23
- Generating the histone code
- When coupled SAHA to
- PIPS (pyrrole-imidazole
- Polyamides) got gene-
- specific DNA binding
- gene activation
24
Generating the histone code Other drugs are
activators of histone deacetylases Resveratrol
increases Sirtuin 1 expression and activity,
possibly by enhancing its binding to Lamin A