Population genetics - PowerPoint PPT Presentation
1 / 21
Title:
Population genetics
Description:
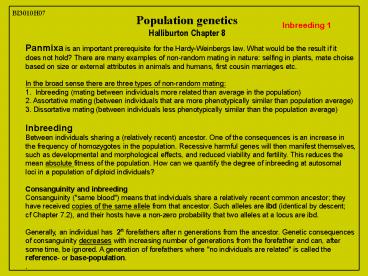
BI3010H07 Population genetics Halliburton Chapter 8 Inbreeding * Panmixa is an important prerequisite for the Hardy-Weinbergs law. What would be the result if it – PowerPoint PPT presentation
Number of Views:110
Avg rating:3.0/5.0
Title: Population genetics
1
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 1
Panmixa is an important prerequisite for the
Hardy-Weinbergs law. What would be the result if
it does not hold? There are many examples of
non-random mating in nature selfing in plants,
mate choise based on size or external attributes
in animals and humans, first cousin marriages
etc. In the broad sense there are three types
of non-random mating 1. Inbreeding (mating
between individuals more related than average in
the population) 2. Assortative mating (between
individuals that are more phenotypically similar
than population average) 3. Dissortative mating
(between individuals less phenotypically similar
than the population average) Inbreeding Between
individuals sharing a (relatively recent)
ancestor. One of the consequences is an increase
in the frequency of homozygotes in the
population. Recessive harmful genes will then
manifest themselves, such as developmental and
morphological effects, and reduced viability and
fertility. This reduces the mean absolute
fitness of the population. How can we quantify
the degree of inbreeding at autosomal loci in a
population of diploid individuals? Consanguinity
and inbreeding Consanguinity ("same blood") means
that individuals share a relatively recent common
ancestor they have received copies of the same
allele from that ancestor. Such alleles are ibd
(identical by descent cf Chapter 7.2), and
their hosts have a non-zero probability that two
alleles at a locus are ibd. Generally, an
individual has 2n forefathers after n
generations from the ancestor. Genetic
consequences of consanguinity decreases with
increasing number of generations from the
forefather and can, after some time, be ignored.
A generation of forefathers where "no individuals
are related" is called the reference- or
base-population. .
2
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 2
Various ways of estimating the degree of
relatedness between two individuals are (CR)
Coefficient of relationship the expected
proportion of the alleles that are ibd. In a
group of offspring (full-sibs) from non-related
parents the proportion is ½ (i.e. half of the
alleles are ibd). (CC) Coefficient of
consanguinity (after Malecót) is a more useful
measure it is the probability that two alleles,
each drawn randomly from the same locus, are ibd.
This measure is identical to the coefficient of
coancestry and coefficient of kinship. To sum
up the probability that two individuals have
received the same allele is the coefficient of
relationship (CR), while the probability of
drawing just that allele from the population gene
pool is the coefficient of coancestry (CC), which
is exactly half the CR and is often referred to
as g. Coeff. of consanguinity (g) coeff. of
kinship coeff. of coancestry ½ coeff. of
relationship gxy S 1/2
Pr(XAi) x 1/2Pr(YAi) (summed over alle
alleles at a locus)
We will hereafter call g the coefficient of
coancestry "Path analysis" for the coefficient
of coancenstry (g). (Fig. 8.1 in
Halliburton) For two fullsibs, in total 4
alleles in two individuals g 4 ½(½) x (½(½)
41/16 1/4 For first cousins, in total 4
alleles in two individuals g 4 ½(¼) x (½(¼)
41/64 1/16
3
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 3
The inbreeding coefficient We now define the
inbreeding coefficient as the probability that an
individual has two alleles that are ibd at a
locus. Because the alleles of an individual are
randomly sampled (half from each of its
parents), its inbreeding coefficient (f) is the
same as the coancestry coefficient of its
parents, i.e. g. The symbol used for the
coefficient of inbreeding is f, with a subscript
which indicates which of the individuals in the
pedigree are involved (see Fig. 8.3 page 275 in
Halliburton). Again The inbreeding coefficient
(f) of an individual the "coancestry
coefficient" (g) of its parents By "path
analysis" it is fairly easy to find the
inbreeding coefficient when the pedigree is
known. (see examples on the following pages).
4
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 4
5
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 5
NB! Table 8.1 contains an error in the "path" for
parent B (K should be F).
6
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 6
The effect of inbreeding on heterozygosity Inbree
ding increases the frequency of homozygotes and
reduces the frequency of heterozygotes in a
population, compared to the reference population
in a former generation. NB! In an inbred
population the homozygosity is caused both by
ibs alleles from the reference population and
ibd alleles due to inbreeding since the
reference population. If we let the subscripts
r and f refer to the reference (base)
population and the inbred population,
respectively, the following is valid Hf Hr
(1-f), which means that f can be interpreted as
a measure of the proportional reduction in the
frequency of heterozygotes relative to the
reference population. (cf expression 8.4 page
275 in Halliburton). Recapitulate
expression 3.19 page 81 in Halliburton, and see
that if no other forces than inbreeding affect
genotype proportions in the population, the
inbreeding coefficient can be expressed as f
(Hexp - Hobs) / (Hexp) cf. expr.
(8.7) NB! Reduction in heterozygosity does not
affect allele frequencies (cf page 278).
7
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 7
Summing up At the individual level f is the
probability that two alleles are ibd. At the
population level f is the proportional reduction
in heterozygosity in an inbred population
relative to an non-inbred reference (base)
population. If no other evolutionary processes
are are in action, these two meanings
are equivalent.
8
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 8
Inbreeding in small populations Small
populations have a non-zero probability that two
alleles are ibd even under panmixia. In the first
generation after the reference population this
probability is 1/(2N) (Halliburton Chapter 7.2
and Fig. 8.4)), and it will increase each
generation so that ft1 1/(2N) (1 - 1/(2N))
ft
(Box 8.1) if the reference
population itself was not inbred, then ft 1 -
(1 - 1/(2N))t and the heterozygosity ... Ht
H0(1 - 1/(2N))t
(NB! Important formula!)
9
BI3010H06
BI3010H06
Inbreeding 9
Population genetics Halliburton Chapter 8
Selfing With selfing, the heterozygosity is
reduced by 50 each generation, and rapidly
approaches zero. After only 10 generations it is
practically zero. In many plants, selfing is not
obligatory, and they may maintain a
certain (although low) heterozygosity (e.g. H
0.00024 in Arabidopsis thaliana).
Repeated full-sib mating Ht1 ½ Ht ¼ Ht-1
(Halliburton Expression (8.22) and Fig. 8.10)
For other types of repeated inbreeding mating
systems, see Halliburton Table 8.4 page 287).
10
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 10
Inbreeding depression Also at single-locus
traits the result of inbreeding is an increased
frequency of homozygotes, and thereby inreased
frequency of harmful, recessive alleles in double
dose so that they manifest in disease/death. This
has been thoroughly documented in studies on
offspring from related parents. For multilocus
traits (quantitative traits) the manifested
effects are quite diverse, like higher
frequencies of harmful morphological deformities,
miscarriages, infant deaths, and mental
retardations in man. In captive animals
(livestocks and pets), typical effects can also
relate to health, longeivity, fertility, general
vigour, heart disease, egg shell thickness
(poultry, fish) and hip dysplasia (e.g. dogs).
At the population level, Frankham (1998) showed
that small, isolated island populations increased
their probability of extinction when the
inbreeding coefficient increased beyond 0.5. He
also showed that such values are quite common in
many small populations.
11
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 11
Inbreeding and "purging" of harmful alleles It
has been suggested that inbreeding, because of
the increased homo-zygosity with harmful alleles
in double doses and lower frequencies of
heterozygotes, can serve a useful cleansing of
the genepool. It gives selection the chance to
get rid of harmful alleles in a process called
purging. Even if it is in principle possible,
opinions are divided as to how efficient his
process can be in natural populations and
domesticated brood stocks. Outbreeding, hybrid
vigour, and outbreeding depression. Outbreeding
is the opposite of inbreeding i.e. mating
between individuals less related than the average
in the population. The outbred population can
have higher fitness than any of the involved
inbred populations because of so-called "hybrid
vigour". However, if local populations have been
adapting to their milieu in many generations and
so-called co-adapted gene complexes have been
formed, these complexes can be broken up by
outbreeding and result in so-called otbreeding
depression. (This has been suggested as a threath
to Norwegian wild salmon stocks under the
influence of escapees from the salmon farming
plants).
12
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 12
Assortative and disassortativ mating
(non-panmixia) Assortative Mating between
individuals that are (phenotypically) more
similar than the average in the population
("alike seeks alike"). This will reduce the
population's heterozygosity for the trait.
Assortative mating may play an important role in
speciation processes (e.g. for sympatric
speciation page 301 ff). (Speciation Read
about pre- and post-mating isolation mechanisms,
sympatric and allopatric speciation on page 301
ff in Halliburton). Disassortative Mating
between individuals that are phenotypically less
alike than the average in the population.
"Contrasts attract each other"). In this case the
heterozygosity increases compared to a panmictic
scenario. This phenomenon is strongly associated
with selection (incl. sexual selection).
13
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 13
Inbreeding and Gametic (genetic)
disequilibrium In section 4.2, the coefficient
of gametic disequilibrium was defined as D
g1g4 - g2g3 Where the gs are the frequencies of
the two-locus gamete types. The disequilibrium
decays over generations (if no selection) at a
rate that depends on the recombination rate r.
The recursion equiation is Dt1
(1-r)Dt Since recombinations only occur in
double heterozygotes, and inbreeding reduces
heterozygosity at all loci, the frequency of
double heterozygotes would expected to be lower
under strong inbreeding (e.g. under selfing).
Hence initial gametic combinations will rarely be
broken up, and decay to gametic equilibrium will
be much slower than in a panmictic population. We
should therefore expect to find high levels of D
in predominantly self-fertilizing species. This
has been confirmed in many studies. (cf page 294
ff).
14
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 14
15
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 15
16
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 16
17
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 17
18
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 18
19
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 19
20
BI3010H07
Population genetics Halliburton Chapter 8
Inbreeding 20
21
(No Transcript)