Diapositiva 1
Title: Diapositiva 1
1
2
Uracil, U Uridine, U Uridine monophosphateUMP
3
Cytosine, C Cytidine, A Cytidine monophosphateCMP
Cytidine, A Cytidine monophosphateCMP
Uracil, U Uridine, U Uridine monophosphateUMP
Thymine, T Thymidine, T Thymidine monophosphateTMP
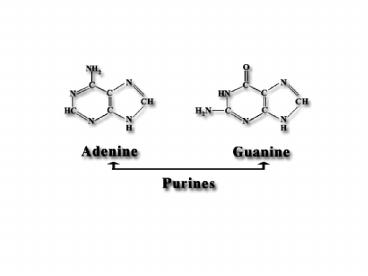
Adenine, A Adenosine, A Adenosine monophosphateAMP
Guanine, G Guanosine, A Guanosine monophosphateGMP
4
syn-Adenosine anti-Adenosine
5
6
A-T Base Pair G-C Base Pair
The double helix of DNA has been shown to exist
in several different forms, depending upon
sequence content and ionic conditions of crystal
preparation. The B-form of DNA prevails under
physiological conditions of low ionic strength
and a high degree of hydration. Regions of the
helix that are rich in pCpG dinucleotides can
exist in a novel left-handed helical conformation
termed Z-DNA. This conformation results from a
180 degree change in the orientation of the bases
relative to that of the more common A- and B-DNA.
Watson Crick original paper
7
(No Transcript)
8
Structure of B-DNA Structure of Z-DNA
9
Parameters of Major DNA Helices
Parameters A Form B Form Z-Form
Direction of helical rotation Right Right Left
Residues per turn of helix 11 10 12 base pairs
Rotation of helix per residue (in degrees) 33 36 -30
Base tilt relative to helix axis (in degrees) 20 6 7
Major groove narrow and deep wide and deep Flat
Minor groove wide and shallow narrow and deep narrow and deep
Orientation of N-glycosidic Bond Anti Anti Anti for Py, Syn for Pu
Comments most prevalent within cells occurs in stretches of alternating purine-pyrimidine base pairs
10
Thermal Properties of DNA As cells divide it is a
necessity that the DNA be copied (replicated), in
such a way that each daughter cell acquires the
same amount of genetic material. In order for
this process to proceed the two strands of the
helix must first be separated, in a process
termed denaturation. This process can also be
carried out in vitro. If a solution of DNA is
subjected to high temperature, the H-bonds
between bases become unstable and the strands of
the helix separate in a process of thermal
denaturation. The base composition of DNA varies
widely from molecule to molecule and even within
different regions of the same molecule. Regions
of the duplex that have predominantly A-T
base-pairs will be less thermally stable than
those rich in G-C base-pairs. In the process of
thermal denaturation, a point is reached at which
50 of the DNA molecule exists as single strands.
This point is the melting temperature (TM), and
is characteristic of the base composition of that
DNA molecule. The TM depends upon several factors
in addition to the base composition. These
include the chemical nature of the solvent and
the identities and concentrations of ions in the
solution. When thermally melted DNA is cooled,
the complementary strands will again re-form the
correct base pairs, in a process is termed
annealing or hybridization. The rate of annealing
is dependent upon the nucleotide sequence of the
two strands of DNA.
11
J Bacteriol. 1970 March 101(3) 738754.
Reexamination of the Association Between Melting
Point, Buoyant Density, and Chemical Base
Composition of Deoxyribonucleic Acid J. De
Ley Laboratory for Microbiology and Microbial
Genetics, Faculty of Sciences, State University,
Ghent, Belgium Abstract The equations currently
used for the calculation of the chemical base
composition of deoxyribonucleic acid (DNA),
expressed as moles per cent guanine plus cytosine
( GC), from either buoyant density (?) or
midpoint of thermal denaturation (Tm) were
recalculated by using only sets of data on DNA
determined with the same strains. All available
information from the literature was screened and
supplemented by unpublished data. The results
were calculated by regression and correlation
analysis and treated statistically. From the data
on 96 strains of bacteria, it was calculated
that GC 2.44 (Tm 69.4). Tm appears to be
unaffected by the substitution of cytosine by
hydroxymethylcytosine. This equation is also
valid for nonbacterial DNA. From the data on 84
strains of bacteria, the relation GC 1038.47
(1.6616) was calculated. The constants in this
equation are slightly modified when data on
nonbacterial DNA are included. Both correlations
differ only slightly from those currently used,
but now they lean on a statistically sound basis.
As a control, the relation between ? and Tm was
calculated from data of 197 strains it agrees
excellently with the above two equations.
pdf de larticle
12
(No Transcript)