Bi430/530 - PowerPoint PPT Presentation
Title:
Bi430/530
Description:
Bi430/530 Theory of Recombinant DNA Techniques First part of course: Technical aspects of molecular biology work--Molecular Cloning Second part of course: – PowerPoint PPT presentation
Number of Views:218
Avg rating:3.0/5.0
Title: Bi430/530
1
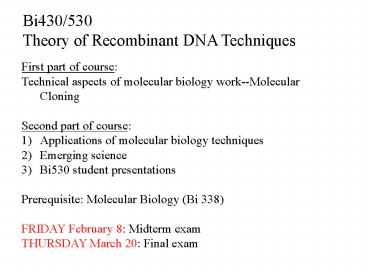
Bi430/530 Theory of Recombinant DNA Techniques
- First part of course
- Technical aspects of molecular biology
work--Molecular Cloning - Second part of course
- Applications of molecular biology techniques
- Emerging science
- Bi530 student presentations
- Prerequisite Molecular Biology (Bi 338)
- FRIDAY February 8 Midterm exam
- THURSDAY March 20 Final exam
2
Syllabus
3
Syllabus--first half
The basics of DNA manipulation (and the Molecular
Cloning Manual)
4
Molecular Cloning (2001), Sambrook and Russel
(3rd ed.)
See also Course Reading 2 (detailed table of
contents) in course packet
5
Syllabus--second half
Using DNA manipulation techniques
6
(No Transcript)
7
(No Transcript)
8
Recombinant DNA
9
Information flow in the cell
DNA
RNA
protein
10
Evolution a dialog between the genome and its
environment
Replication/mutation
DNA
RNA
protein
Natural selection
11
Organisms respond to their environment via
information from sensory input and changes in
gene expression
DNA
RNA
protein
environment
Dynamic, immediate, transient modification of DNA
program
12
Species respond to environment over long time
frames via mutations in the DNA program
DNA
RNA
protein
evolution
environment
--Stable (permanent) --reflects effect of
environment over large time scales
13
Human activity transient modifications of
environment, permanent modifications of DNA
program
DNA
RNA
protein
Human intervention genetics (indirect), rDNA
(direct)
environment
14
2006 53 years of DNA structure
Rosalind Franklin and Maurice Wilkins X-Ray
fiber diffraction pattern of pure B-form DNA
(1953) James Watson and Francis Crick Proposed
two antiparallel, helical strands forming a
stable duplex with DNA bases on interior of the
molecule, joined by hydrogen bonds (1953) But
DNA was not discovered in 1953--it had been known
as the element of genetic transmission at least
since 1947, when Avery showed that DNA could
transform bacterial colony morphology Why was
the structure so important?
15
Structure of DNA
To Watson and Crick, the structure
suggested --Mechanism for replication --Stability
for information storage, yet accessing the
information not difficult The DNA structure
provided a new template for hypotheses regarding
biological phenomena (amenability to study)
16
DNA is easy to work with
- Readily isolated--plasmid isolation, PCR
- Stable--not chemically reactive like RNA (even
archaeologically stable!) - Easy to propagate and move from cell to cell
- Easy to make specific constructs
- Easy to make specific mutations
- Very easy to sequence (record-keeping)
- Predictable behavior
- Sequence lends itself to analysis--genome projects
17
DNA is very easy to sequence
Etc.
18
http//www.genomesonline.org/
19
The behavior of DNA (genes) is predictable
Gene sequence conservation often
indicates functional similarity Non-protein
coding information sequences can often be
detected by homology (promoters for transcription
initiation, transcription terminators, ribosome
binding sites, DNA binding protein binding sites)
the genetic code
20
The genetic code and the roots of biotechnology
1961 Marshall Nirenberg and Heinrich J.
Matthaei polyU mRNA encodes poly-phenylalanine 1
966 Nirenberg and colleagues had deciphered the
61 codons (and 3 nonsense codons) for all 20
amino acids 1968 Nobel prize for Nirenberg,
Holley, and Khorana
21
1966 George and Muriel Beadle write The
deciphering of the DNA code has revealed our
possession of a language much older than
hieroglyphics, a language as old as life itself,
a language that is the most living language of
all--even if its letters are invisible and its
words are buried deep in the cells of our bodies.
22
The public reaction to the deciphering of the
genetic code Wow just as big a breakthrough
in biology as Newton's discovery of gravitation
in the seventeenth century was in physics.
--John Pfeiffer, journalist, 1961 Optimism No
stronger proof of the universality of all life
has been developed since Charles Darwin's 'The
Origin of Species' demonstrated that all life is
descended from one beginning. In the far future,
the hope is that the hereditary lineup will be so
well known that science may deal with the
aberrations of DNA arrangements that produce
cancer, aging, and other weaknesses of the
flesh. Chicago Sun-Times, 1962
23
Caution knowledge gained from the genetic code
might well lead in the foreseeable future to a
means of directing mutations and changing genes
at will. 1961, A. G. Steinberg of Case Western
Reserve University knowledge of the genetic
code could lead to methods of tampering with
life, of creating new diseases, of controlling
minds, of influencing heredity, even perhaps in
certain desired directions.1961, Arne Wilhelm
Kaurin Tiselius, 1948 Nobel Laureate in Chemistry
24
1967 Will Society Be Prepared? Marshall
Nirenberg, editorial in Science (see WebCT)
25
Nirenberg, 1967
"When man becomes capable of instructing his own
cells, he must refrain from doing so until he has
sufficient wisdom to use this knowledge for the
benefit of mankind.... Decisions concerning
the application of this knowledge must ultimately
be made by society, and only an informed society
can make such decisions wisely."
26
Response from Joshua Lederberg, 1967 (see letter
on WebCT) (paraphrased) -- We need to be
particularly careful with manipulation of the
germ cell lines (heritable changes). --
Considerations governing control of our biology
are equally important to considerations governing
control of our cultural institutions (given that
culture is mutable and heritable)
27
1975 The Asilomar Conference on Recombinant DNA
- 1974 moratorium on recombinant DNA research
- new technology created extraordinary novel
avenues for genetics and could ultimately provide
exceptional opportunities for medicine,
agriculture and industry. concerns that
unfettered pursuit of this research might
engender unforeseen and damaging consequences for
human health and the Earth's ecosystems
http//nobelprize.org/nobel_prizes/chemistry/artic
les/berg/index.html
28
1975 The Asilomar Conference on Recombinant DNA
- Conference included internationally prominent
scientists, government officials, doctors,
lawyers, members of the press - Conclusion recombinant DNA research should
proceed but under strict guidelines. - The moratorium was lifted, and guidelines were
subsequently promulgated by the National
Institutes of Health and by comparable bodies in
other countries.
http//nobelprize.org/nobel_prizes/chemistry/artic
les/berg/index.html
29
- The Asilomar principles
- containment should be made an essential
consideration in the experimental design - the effectiveness of the containment should match
the estimated risk as closely as possible. - Additional suggestions
- Use biological barriers to limit the spread of
recombinant DNA - fastidious bacterial hosts that are unable to
survive in natural environments - nontransmissible and equally fastidious vectors
(plasmids, bacteriophages, or other viruses) that
are able to grow in only specified hosts
30
- The Asilomar principles
- Safety factors
- physical containment, exemplified by the use of
hoods or where applicable, limited access or
negative pressure laboratories - strict adherence to good microbiological
practices, which would limit the escape of
organisms from the experimental situation - education and training of all personnel involved
in the experiments would be essential to
effective containment measures.
31
- Regulation of biotechnology US National
Institutes of Health (NIH) Guidelines - stipulations of biosafety and containment
measures for recombinant DNA research - delineations of critical ethical principles and
safety reporting requirements for human gene
transfer research - See http//www4.od.nih.gov/oba/Rdna.htm
- (see also CR 3 in course packet)
32
Unnatural Selection (first reading in the
course packet) By Allison Snow Is the process
for altering genes (evolution vs. human)
irrelevant? Product (transgenic organism) is
the phenotype the only thing that is
important? Reverberations from introduction of
modified organisms? Spread of gene? Effects of
spread? Success of organism? Effects on other
organisms? The technologys main hazards are
probably yet to manifest themselves
33
What can we do with recombinant DNA technology?
- begin to learn how cells, tissues, organisms,
communities work, interact, respond to the
environment (gain scientific knowledge) - improve human health
- industrial production of useful enzymes,
metabolic products - improve industrial process
- raise agricultural productivity
- investigate problems of geneology, paternity,
anthropology, archaeology - investigate criminal cases
- etc.
34
How is recombinant DNA technology useful in
medicine?
- Diagnosis of disease
- Animal models for human diseases
- Therapies
- nucleic acids gene therapy
- pharmacologically active proteins
- small molecule design and testing
- Antimicrobials
- Vaccines
- Microbicides
35
The biotechnology industry is very new Case in
point Genentech (S. San Francisco)
2006 Genentech to open production facility in
Portland (2010)
36
- Day 1 summary
- The simplicity of a DNA-based information system
makes genetic manipulation possible - This represents an unprecedented level of
interaction with living systems - Benefits and costs of technology require
continuous assessment
37
Visualizing DNA (and RNA, protein) non-specific
detection methods
- Quantitation of DNA (Course Reading 4)
- Electrophoresis (Course Reading 5 )
- Visualizing DNA ( protein) in gels (Course
Reading 6)
38
Quantitation of DNA by UV absorbance
- Measure absorbance of UV light by sample (the
aromatic bases have a characteristic absorbance
maximum at around 260 nanometers) - 1.0 A260 (1 cm light path) DNA concentration
of 50 micrograms per ml (double stranded DNA) or
38 micrograms per ml (single-stranded DNA or RNA) - the effective range for accurate measurement is
rather narrow A260 from 0.05 to 2.0 (DNA
concentrations from 2.5 to 100 micrograms/ml) - Sample must be very pure for accurate
measurements (RNA, EDTA and phenol all absorb at
260 nm)
39
How can concentration be determined by
absorbance? DNA has a characteristic molar
extinction coefficient ? The Beer-Lambert
law I Io10- ?dc I intensity of
transmitted light Io intensity of incident
light ? molar extinction coefficient d
optical path length c concentration of
absorbing material
How much light gets through a solution depends on
whats in it and how much of it there is
40
- The Beer-Lambert law
- I Io10- ?dc
- Absorbance A measured by a spec is log I/Io
- When path length d 1 cm, A is called the
optical density OD - If you know the ?, the absorbance of a solution
will tell you the concentration - OD? ?c
- for nucleic acids
- dsDNA 6.6
- ssDNA, RNA 7.4
- (but these values change with pH and salt
concentration!)
41
A typical (good) scan (multiple wavelengths) of
a DNA sample
A260/A280 1.8 is good (lower values indicate
significant protein contamination)
0.5
A260 0.327
Absorbance (1 cm path length)
0
200
260
400
Wavelength (nm)
42
How does A260 give you the quantity of DNA?
Example sample of 250 base pair fragment of DNA
has an A260 .327 What is its molar
concentration? Given (1.0 A 50 micrograms/ml
DNA) DNA conc. .327 x 50 16.35
micrograms/ml MW of an average bp. 650
Daltons Therefore 250 bp. Fragment has a MW of
1.6 x 10 5 Daltons Solve for molarity 1.02 x 10
-7 M, or 102 nanomolar (nM) Important to know
how to do this calculation
43
What is the molarity of a 16.35 microgram/ml
solution of a 250 base pair DNA fragment?
1 gram
1 mole
1000 ml
16.35 micrograms
1 ml
106 micrograms
1 L
1.6 x 105 grams
1.02 x 10 -7 molar 0.102 x 10-6 molar 0.1
micromolar (?M) 102 x 10-9 molar 102 nanomolar
(nM)
44
- Fluorometry another method for quantitation of
DNA - Hoechst 33258 (a fluorescent dye)
- Binds to DNA in the minor groove (without
intercalation) - Fluorescence increases following binding
- Good for quantitation of low concentrations of
DNA (10-250 ng/ml) - rRNA and protein do not interfere
- But you need a fluorometer
45
- Another method for quantitation of DNA
- Ethidium bromide (fluorescent dye) binding
- Compare sample DNA fluorescence to standards of
known concentration (dilution series) - In solution or using gel electrophoresis
A commercially available quantitative DNA standard
46
Visualizing DNA Electrophoresis
- Allows separation of biomolecules (DNA, RNA,
protein) on basis of size - A separation matrix, or gel (agarose or
polyacrylamide), is saturated with an
electrically conductive buffer - Samples are loaded, an electric field is applied,
and negatively charged biomolecules in the sample
travel toward the cathode - The larger the molecule, the slower the travel
through the gel matrix - Dyes allow a visual estimate of the rate of
travel through the gel - The choice of matrix depends mainly on the size
of DNA being analyzed
47
Agarose gels
- Agarose a polysaccharide polymer of alternating
D- and L-galactose monomers, isolated from
seaweed - Pore size is defined by the agarose concentration
(higher concentration, slower DNA migration
overall) - The conformation of the DNA (supercoiled, nicked
circles, linear) affects the mobility of the DNA
in gels - Rate of DNA migration is affected by voltage (5
to 8 Volts/cm is close to optimal) - Agarose comes in a myriad of types (variable
melting temperatures, generated by differential
hydroxyethylation of the agarose)
48
Agarose gels
- Standard gels can separate DNA fragments from 100
bp to about 20,000 bp - Pulsed-field gels separate very large DNA
fragments (up to 10,000,000 bp, or 10 Mb)
This apparatus allows periodic shifts in the
direction of DNA migration 120 refers to the
reorientation angle (difference between
orientation of electric fields A and B
49
Typical agarose gel
Load samples in wells
xylene cyanol
bromophenol blue
-
?
(the DNA fragments are not visible without some
sort of staining)
time of electrophoresis (progress monitored by
marker dyes)
50
(No Transcript)
51
Polyacrylamide gels
- Acrylamide monomers (toxic!) polymerized to form
gel matrix - The gel structure is held together by the
cross-linker-- usually N, N'-methylenebisacrylamid
e ("bis" for short) - Pore size defined by concentration of gel (total
percentage) and concentration of the crosslinker
(bis) relative to acrylamide monomer - Very high resolution (better than agarose)
- Suitable for separation of nucleic acids from 6
to 1000 base pairs in length
52
Polyacrylamide gels
- Native gels (DNA stays double-stranded)
- Denaturing gels--run in the presence of high
concentrations of denaturant (usually urea) and
at high temperature DNA is single stranded
(sequencing gels) - (also useful in separation of proteins, when
proteins are treated with SDS, which denatures
proteins and gives a uniformly negative surface
charge)
53
Recipe for a polyacrylamide gel
- Acrylamide (anywhere from 4 to 20 , depending
size of nucleic acids or proteins in the gel) - Bis-acrylamide (the ratio of Bis to regular
acrylamide is important) - Water
- Buffer
- To initiate polymerization, add
- APS Ammonium persulfate
- -- generates free radicals needed for
polymerization - TEMED N,N,N,N - tetramethylethylenediamine
- -- accelerates free radical generation by APS
54
- More about gels
- There has to be a buffer (for carrying current)
- TAE (Tris-acetate-EDTA) good resolution of DNA,
but buffering capacity is quickly depleted - TBE (Tris-borate-EDTA) High buffering capacity,
resolution is pretty good - Use gel loading buffers (relatively simple)
- Dense material to carry sample to bottom of wells
(sucrose, glycerol, or ficoll) - Dyes for tracking progress of electrophoresis
- Bromophenol blue fast migration
- Xylene cyanol slow migration
- Occasionally denaturant is present (formamide)
for denaturing gels (e.g. sequencing gels)
55
- Protein electrophoresis
- Almost always polyacrylamide based
- The anionic detergent SDS (sodium dodecyl
sulfate) is used to denature the proteins, giving
each protein a uniform negative charge - Protein separation occurs as a function of size
- Discontinous Tris-Cl/glycine buffer system
- Stacking gel pH 6.8, low polyacrylamide
concentration, focuses proteins into thin layer
(gives higher resolution upon separation) - Separating gel pH 8.8, separates proteins on the
basis of size
56
Polyacrylamide gel set up (protein gels)
Stacking gel at low pH, glycine is protonated
(no neg. charge), Cl- ions at the leading edge,
glycine trailing, steep voltage gradient in
between, thats where the proteins get focused
into a thin band Separating gel at higher pH,
glycine deprotonates, runs with the Cl- at the
leading edge, and the proteins separate based on
size
57
Staining nucleic acids
- ethidium bromide, an anti-trypanosomal drug for
cattle - Stain works by intercalating in stacked base
pairs, elongates DNA helix - Fluorescence increases upon DNA binding
- Stained bands visualized by UV illumination (302
or 260 nm)
G-C base pair
Ethidium bromide
58
Example of an agarose-DNA gel, Stained with
ethidium bromide
Fragments of bacteriophage ? genomic DNA (48 kb)
cut with the restriction enzyme Hind III The
fragments are equimolar--why is the band
intensity different?
Direction of electrophoresis
59
Another ethidium bromide-stained agarose gel
samples
M
The marker lane (M) gives size standards for
comparison with the sample lanes
60
Other methods for staining DNA
- SYBR gold (Molecular Probes, Eugene, OR), more
than 10-fold more sensitive than ethidium bromide
for detecting DNA, but expensive! - methylene blue not toxic, but the staining
protocol is time consuming, and sensitivity
somewhat lower than ethidium bromide - silver staining high degree of sensitivity, but
the protocols are time consuming, and proteins
are also stained by silver
61
Protein detection in gels
- Coomassie Brilliant Blue R-250 dye from the
textile industry that has a high affinity for
proteins - Proteins in gels must be fixed (rendered
insoluble) first with acetic acid/methanol - Dye probably interacts with NH3- groups of the
proteins, also through van der Waals forces
http//www.galab.de/laboratories/services/biopharm
a/img/sds.jpg
62
Protein detection in gels
- Silver staining
- 100 to 1000-fold more sensitive than Coomassie
stain for detecting proteins (need far less
sample to see it on a gel) - Process relies on differential reduction of
silver ions bound to amino acid side chains (like
the photographic process) - There is protein-to-protein variability of
staining
ugly (keratin)
good
bad (overstained)
63
Protein detection in gels
- Sypro Ruby (Molecular Probes inc, proprietary
compound) - As sensitive as silver staining, less variability
- Fast protocol
- Expensive
1 nanogram of protein
Standard gel
2-D gel
64
Visualizing DNA (and RNA, protein) non-specific
methods
- Quantitation of DNA (Course Reading 4)
- Electrophoresis (Course Reading 5 )
- Visualizing DNA ( protein) in gels (Course
Reading 6)
65
Methods for detecting specific biomolecules
- Southern blots (DNA-DNA hybridization)(Methods
for labeling probe DNA) CR7, MC 6.33 - 6.38 - B. Northern blots (DNA-RNA hybridization) CR8,
- MC 7.21 - 7.26, MC 7.82 - 7.84
- Western blots (detection of proteins with
specific antibodies) CR9, MC A9.28, MC A8.52-A8.55
66
Visualizing DNA, RNA and Protein detecting
specific sequences or proteins
- Techniques allow one to distinguish specific
sequences or proteins in a large, mixed
population, e.g. in cell extracts or genomic DNA
preparations - For DNA and RNA, specific sequence detection is
based on DNA and RNA complementarity and
base-pairing - For proteins, the specific detection is based on
antibodies that recognize the protein of interest
(or based on a specific assay for activity of the
protein)
67
Detecting specific DNA sequences the Southern
blot
68
Immobilization of nucleic acids
nitrocellulose or nylon membrane boundary DNA
binds to it
Agarose or Polyacrylamide gel
A typical capillary blotting apparatus.
Electroblotting is also commonly used
69
Southern blotting Immobilization of target DNA
and detection
- DNA is fixed to the nylon membrane by
- Baking, 80C
- UV crosslinking (links thymines in DNA to
charged amine groups in membrane), DNA only - Probe to detect sequence of interest by
base-pairing (hybridization) - Obtain probe DNA synthetic oligonucleotide or
cloned gene (single stranded) - Label probe for later detection
- Radioactivity
- Non-radioactive label
70
Radioactive probes 32P labeling
- Use T4 polynucleotide kinase
- --catalyzes the transfer of the gamma phosphate
of 32P ATP to the 5 end of DNA fragment to be
used as a probe - 32P is a high energy beta particle emitter, and
provides good sensitivity for detection of
hybridization between the probe DNA and the
target (blot) DNA - Detect radiolabel with
- --autoradiography (X ray film)
- --phosphorimager (phosphor coated plates store
the energy of the radioactive particle, laser
excitation releases photons of light that are
collected and represented as a picture, greater
dynamic range than film, and faster too
71
Non-radioactive labels
e.g. horseradish peroxidase
oxidation
72
Non-radioactive labels
or digoxygenin/antibody-conjugated HRP
oxidation
can also use biotinylated DNA probe
73
Hybridize probes to membranes
- blocking agents (e.g. milk, SDS) prevent
non-specific interactions between probes and
membrane - Volume exclusion agents (eg. dextran sulfate)
increase rate and level of hybridization - Wash blot with increasing stringency
- Low stringency high salt, low temperature, probe
binds to sequences with mismatches - High stringency low salt, higher temp., probe
binds only to fully complementary sequences
74
Southern Blot--one example
(or PCR fragment)
(RFLPs)
75
Northern blots
Same basic technique as Southern blots, but RNA
is run on the initial gel and is transferred to
the membrane. Use this method to measure levels
of gene transcription in vivo (detecting changes
in the levels of RNA transcript under differing
conditions) Microarrays for measuring mRNA
abundance are based on this principle, but many
probes are immobilized in a regular array --
reverse transcribed (and fluorescently labelled)
RNA lights up the probes on the microarray
76
Western blots proteins
Proteins are transferred to membranes using
the same principle as Southern
blots Specific proteins detected by probing blot
with antibodies to protein of interest Antibody
binding is detected by antibody to the original
antibody that has enzyme (horseradish peroxidase,
alkaline phosphatase) or radioactivity (125I)
conjugated to it
77
Methods for detecting specific biomolecules
- Separate DNA, RNA, or proteins on the basis of
size (gel electrophoresis) - Immobilize the separated DNA, RNA, or protein
- Probe the blot with something that will
specifically interact with a target - DNA and RNA complementary nucleic acid
- Protein antibody to that protein
78
(No Transcript)