Enzymes/ Kinetics - PowerPoint PPT Presentation
1 / 83
Title:
Enzymes/ Kinetics
Description:
Enzymes/ Kinetics Study of enzyme catalyzed reactions Rate of reactions Enzyme specificity Mechanism of catalysis There are several good reasons for adding kinetic ... – PowerPoint PPT presentation
Number of Views:239
Avg rating:3.0/5.0
Title: Enzymes/ Kinetics
1
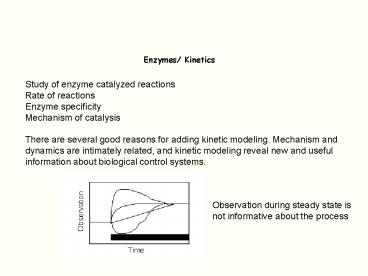
Enzymes/ Kinetics
Study of enzyme catalyzed reactions Rate of
reactions Enzyme specificity Mechanism of
catalysis There are several good reasons for
adding kinetic modeling. Mechanism and dynamics
are intimately related, and kinetic modeling
reveal new and useful information about
biological control systems.
Observation during steady state is not
informative about the process
2
Catalyst
Catalyst Enzyme biocatalyst e.g. Fermentation
of sugar to ethanol by yeast enzymes Conversion
of fatty acids to polyketide antibiotics by
filamentous fungi Conversion of milk to cheese
by microorganisms Conversion of sugar to CO2 by
bakers yeast to make leavened bread
3
BioCatalyst
MOST Enzymes are Proteins Enzyme advantages over
chemical catalysis Enzyme
concentration is very small and enzyme is almost
always limiting
4
Enzyme function
S lt---------gtP 1 2 3 DGo standard free
energy change for chemical reactions DGo
standard free energy change for biological
reactions 298o k (RT) 1 atm, 1M substrate,
pH7.0 Active site- pocket in enzyme that
binds substrates- most complimentary in structure
to transition state
5
Rationale for enzyme kinetics
Conformation of proteins and positions of side
chains are important for enzyme-substrate
interactions and catalysis. Forces involved in
protein folding and structure are also involved
in catalysis- enzyme-substrate specificity
Trypsin hydrolyses proteins by cleaving peptide
bond adjacent to Lys/Arg. Aspartate residue in
trypsin active site mediates ionic interaction
with Lys /Arg and this arranges protein residue
at which hydrolysis occurs. To use enzymes in
biotechnology, pharmaceutics or drugs that
inhibit enzymes in medicine, you NEED TO KNOW
KINETIC PARAMETERS OF THE ENZYME REACTION. We
may want enzymes that WORK FAST- convert more
substrate in a fixed unit of time. To do this
optimization we have to perform and analyze the
enzyme catalyzed reaction. You can adjust pH,
temperature and add co-factors to optimize enzyme
activity. You cannot adjust substrate
selectivity. Just like chemical reactions,
enzyme catalyzed reactions have kinetics and
rates Reaction kinetics is Michaelis-Menten
kinetics.
6
Enzyme-substrate cycle
7
Activation energy
Free energy
Ground state starting point (energetically
speaking) - - - - -
8
Energy Barriers
S
Free energy
P
- - - - - -
9
Sucrose to CO2 and H2O
C12H22O11 12O2 lt-----gt 12CO2 11 H2O Reaction
has large negative DGo Therefore Product
prevails at equilibrium In your pantry you can
store sucrose in the presence of oxygen. It does
not spontaneously convert into CO2 and
water! ACTIVATION energy of this reaction is
HUGE In a cell sucrose is rapidly converted to
CO2 and H2O ENZYMES!!! Activation energy
barriers are required for ordered life!!! Without
barriers molecules would spontaneously
interconvert- there would be no regulation
10
Reaction coordinate
Overall std free energy change (S goes to P)
11
Catalyzed Vs uncatalyzed reaction
12
Rate enhancements
Enzymatic rate enhancements are 103 to 1017 x
13
How do ENZYMES carry out catalysis?
- -
14
(No Transcript)
15
A to B to C
A------------gt B------------gtC
The rate constant for B to C is smaller. B to C
is slow (rate determining) A is rapidly converted
to B but B accumulates because its conversion to
C is slow Reaction is in only one direction
because DG of B is lower than A and C is lower
than B B to C is rate limiting
conc
DG
Rxn coordinate
time
16
A to B to C
A------------gt B------------gtC
The rate constant for A to B is smaller. A to B
is slow (rate determining) A is not rapidly
converted to B because its conversion to B is
slow. B never accumulates because it is rapidly
converted to C Reaction is in only one direction
because DG of B is lower than A and C is lower
than B A to B is rate limiting
conc
DG
Rxn coordinate
time
17
A to B to C
A------------gt B------------gtC lt-----------
The rate constant for A to B is small. The rate
constant for B to A is large. A to B is slow but
B to A is very fast and B to C is kinda fast. A
is not rapidly converted to B. B never
accumulates because it is very rapidly
reconverted to A and C accumulation is not very
rapid because most of B is reconverted back to A
rather than to C Reaction is in two directions
because DG of B is higher than A and C is lower
than B and A
conc
DG
Rxn coordinate
time
18
A to B to C
A------------gt B------------gtC lt-----------
The rate constant for A to B is small. The rate
constant for B to A is large. A to B is slow but
B to A is kinda fast and B to C is very fast. A
is not rapidly converted to B. B never
accumulates because it is very rapidly converted
to C. or reconverted to A. Accumulation of C is
rapid because most of B is converted to C rather
than reconverted to A Reaction is primarily in
one directions because while DG of B is higher
than A, C is lower than B and A A to B is rate
limiting
conc
DG
Rxn coordinate
time
19
Enzyme catalyzed Steady State
Enz Sub lt-------gt Enz-Sub lt-------gt Enz
Prod
S
P
Conc
During steady state formation of ES equals its
breakdown k1ESk-1ESk2ES Km k-1k2/k1
ES
E
Time
Pre steady state
Steady state
20
Rate of a Reaction
What is the rate of a chemical reaction S----gtP -
- - - - AB----.gtP is a bimolecular
reaction (there are two reactants) and this is a
second order reaction - Sometimes second
order reactions appear as first order. E.g. If
conc of B is very large and does not change much
then reaction rate is entirely dependent on conc
of A only
21
Rate of catalysis
Vo For a fixed amount of enzyme, The rate
of catalysis
22
All measurements are done at very high substrate
conc and very low product conc so the reverse
reaction is rare. We can simplify the above
reaction scheme as
23
Rate constants
k1 k2 A -----gt B ------gt
C lt----- k-1 If k-1 is
greater than k2 (B reforms A faster than it forms
C) then there will be a rate-determining
pre-equilibrium and overall rate of formation of
C will depend on ALL THREE RATE
CONSTANTS ((((dC/dt k1k2/k-1A)))) If
k2 is vastly greater than k-1 (B forms C faster
than it reforms A) then we can effectively ignore
the back reaction (B to A) and the only question
then is whether k1 or k2 is rate limiting.
k1
k2
ES
ES
EP
k-1
- Binding of substrate to enzyme is reversible
- Product release from enzyme is instantaneous
k1 is rate constant for formation of ES k-1 is
rate constant for conversion of ES to ES k2 is
rate constant for formation of product kcat
24
ES
- Rate of formation k1ES
- Rate of breakdown k-1ES k2ES
- Assume steady state
- As ES is produced, it reacts
- ES remains constant
- Rate formation rate breakdown
- So, k1ES k-1ES k2ES
- k1ES (k-1 k2)ES
- Rearrange
- ES / ES (k-1 k2) / k1
- Where (k-1 k2) / k1 KM (Michaelis constant)
- Define total enzyme concentration ET E
ES - Substitute for E
- (ET ES)S / ES KM
- Solve for ES
- ES ETS / KM S
25
V0 initial velocity/rate
- ES ETS / KM S
- Since V0 k2ES
- V0 k2ETS / KM S
- Define maximum velocity Vmax
- Occurs at high S
- Enzyme is all in ES form
- ES ET
- Vmax k2ET
- Therefore
- V0 VmaxS / KM S
- Rate of reaction increase with S
- Rate levels off as approach Vmax
- More S than active sites in E
- Adding S has no effect
- At V0 ½ Vmax
- gt S KM
26
Michaelis-Menten
The classic Michaelis-Menten rate
equation The first order rate constant kcat
is k2 Vmax is product of kcat and enzyme
conc Km is mixture of rate constants that
describe formation and dissociation of
enzyme-substrate complexes Km gives a sense of
the affinity of enzyme for substrate When
substrate is gtgtgt Km, the reaction kinetics are
equal to max rate (Vmax). When substrate is ltltlt
Km, the reaction rate is kcat/Km All these
analyzes are done at saturation kinetics Kcat
and Km are PRIMARY INDICATORS of how well an
enzyme will react with a particular substrate.
High kcat fast reactions. low kcat slow
reactions High Km low affinity of enzyme for
substrate. Low Km high affinity of enzyme for
substrate
27
Michaelis-Menton Equation
- -
- -
- V0
- -
- What is the ES?
28
What do we measure?
- - - - ---- -----
ABlt---gtAB
At low S, Km gtgt S At high S, S gtgt Km
29
Initial Velocity
Velocity measured at very beginning of reaction
when very little product is made Initial velocity
is measured at saturation kinetics- at high S,
enzyme is saturated with respect to substrate
Vmax
Vo
Product
Substrate
Time
30
Vmax and Km
S
V0 Vmax
SKm
k-1 k2
Km
k1
When V0Vmax, Km S
Km is unique to each Enzyme and Substrate. It
describes properties of enzyme-substrate
interactions Independent of enzyme conc.
Dependent on temp, pH etc. Vmax is maximal
velocity POSSIBLE. It is directly dependent on
enzyme conc. It is attained when all of the
enzyme binds the substrate. (Since these are
equilibrium reactions enzymes tend towards Vmax
at high substrate conc but Vmax is never
achieved. So it is difficult to measure). When an
enzyme is operating at Vmax, all enzyme is bound
to substrate and adding more substrate will not
change rate of reaction (enzyme is saturated).
(adding more enzyme will change the reaction).
31
Measuring Km and Vmax
1/vo
1/Vmax
1/S
-1/Km
You can use a curve fitting algorithm to
determine Km and Vmax from a V vs S plot (need
a computer) Reaction rates are initial rates
determined when the substrate is in vast excess
and isnt changing much. Alternatively you can
convert the curve to a straight line via a double
reciprocal plot (1/Vmax and 1/S)
32
Reciprocal
It is not easy to extrapolate a hyperbola to its
limiting value (computers can do this) The
Michaelis-Menten equation can be recast into a
linear form
To obtain parameters of interest Reciprocal form
of equation 1 Km 1 1 V Vmax
S Vmax Y m x b
The y-intercept gives the Vmax value and the
slope gives Km/Vmax
Vmax is determined by the point where the line
crosses the 1/Vi 0 axis (so the S is
infinite). Km equals Vmax times the slope of
line. This is easily determined from the
intercept on the X axis.
33
1/Vo
It is difficult to accurately measure Vmax
S
V0 Vmax
SKm
Reciprocal
1
Km
1 S
1 Vma
V0
Vmax
Vmax k2Et k2 is also called Kcat Et is conc
of active sites in enzyme
Km values of enzymes range from 10-1M to 10-7M
for their substrates. It varies depending on
substrate, pH, temp, ionic strength etc. Kcat is
turnover number for the enzyme-number of
substrate molecules converted into product per
unit time by that enzyme
34
Km and Vmax
Km is S at 1/2 Vmax It is a constant for a
given enzyme at a particular temp and pressure It
is an estimate of equilibrium constant for
substrate binding to enzyme Small Km tight
binding, large Kmweak binding It is a measure of
substrate concentration required for effective
catalysis Vmax is THEORETICAL MAXIMAL
VELOCITY Vmax is constant for a given enzyme To
reach Vmax, ALL enzyme molecules have to be bound
by substrate Kcat is a measure of catalytic
activity- direct measure of production of product
under saturating conditions. Kcat is turnover
number- number of substrate molecules converted
to product per enzyme molecule per unit
time Catalytic efficiency kcat/km Allows
comparison of effectiveness of an enzyme for
different substrates
35
Enzyme Km examples
Hexokinase prefers glucose as a substrate over
ATP
36
Kcat
Catalase is very efficient-it generates 40
million molecules of product per second. Fumarase
is not efficient-it generates only 800
molecules/per second
- kcat Vmax / ET
- Turnover number
- Number of reaction processes each active site
catalyzes per unit time - Measure of how quickly an enzyme can catalyze a
specific reaction - For M-M systems kcat k2
37
Kcat/Km
- Rate constant of rxn E S ---gt E P
- Specificity constant
- Gauge of catalytic efficiency
- Catalytic perfection 108 -109 M-1 s-1 (close to
diffusion)
38
(No Transcript)
39
Enzyme cofactors
40
Coenzymes
41
(No Transcript)
42
How do ENZYMES carry out catalysis?
- Entorpy reduction- holds substrates in proper
position - Bringing two reactants in close proximity (reduce
entropy increase effective reactant concentrat) - Substrate is desatbilized when bound to enzyme
favoring reaction-(change of solvent,
charge-charge interactions strain on chemical
bonds). - Desolvation of substrate- H bonds with water are
replaced by H bonds with active site - Enzymes form a covalent bond with substrate which
stabilizes ES complex (Transition state is
stabilized) - Enzyme also interacts non-covalently via MANY
weak interactions - Bond formation also provides selectivity and
specificity - (H bonds- substrates that lack appropriate groups
cannot form H bonds and will be poor substrates) - (Multiple weak interactions between enzyme and
substrate) - Free energy released by forming bonds is used to
activate substrate (decrease energy barrier/lower
activation energy of reaction) - Induced fit-binding contributes to conformation
change in enzyme - Whats the Bill?
- 5.7 kJ/mol is needed to achieve a 10x increase in
rate of a reaction - Typical weak interactions are 4-30 kJ/mol
- Typical binding event yields 60-100 kJ/mol
43
A Hypothetical reaction
44
Breaking a stick
Imagine you have to break a stick. You hold the
two ends of the stick together and apply force.
The stick bends and finally breaks. You are the
catalyst. The force you are applying helps
overcome the barrier.
45
A stickase with a pocket complementary in
structure to the stick (the substrate) stabilizes
the substrate. Bending is impeded by the
attraction between stick and stickase.
46
An enzyme with a pocket complementary to the
reaction transition state helps to destabilize
the stick, contributing to catalysis of the
reaction. The binding energy of the interactions
between stickase and stick compensates for the
energy required to bend the stick.
47
Role of binding energy in catalysis. The system
must acquire an amount of energy equivalent to
the amount by which DG is lowered. Much of this
energy comes from binding energy (DGB)
contributed by formation of weak noncovalent
interactions between substrate and enzyme in the
transition state.
48
Lock/Key or Induced Fit
49
Lock/Key- Complementary shape
The enzyme dihydrofolate reductase with its
substrate NADP NADP binds to a pocket that is
complementary to it in shape and ionic
properties, an illustration of "lock and key"
hypothesis of enzyme action. In reality, the
complementarity between protein and ligand (in
this case substrate) is rarely perfect,
50
Induced Fit
Hexokinase has a U-shaped structure (PDB ID
2YHX). The ends pinch toward each other in a
conformational change induced by binding of
D-glucose (red).
51
Substrate specificity
The specific attachment of a prochiral center (C)
to an enzyme binding site permits enzyme to
differentiate between prochiral grps
52
Enzyme-substrate
53
Catalysis
Acid-Base Catalysis- donate or accept
protons/electrons from and to substrate Covalent
Catalysis-transient covalent link between
substrate and enzyme side chain Metal-Ion
Catalysis-Metal in active site donate or accept
protons with substrate Proximity Orientation
Effects (reduction in entropy-two mol brought
together and oriented in specific
manner) Transition State Preferential Binding
54
R Groups
- The active sites of enzymes contain amino acid R
groups. - Active site is lined with hydrophobic residues
- Polar amino acid residues in active site are
ionizable and participate in the reaction.
Anion/cation of some amino acids are involved in
catalysis
Lysozyme Cleaves glycosidic bonds in
carbohydrates
55
Covalent Catalysis
All or part of a substrate is transiently
covalently bound to the enzyme to form a reactive
intermediate Group X can be transferred from A-X
to B in two steps via the covalent ES complex
-EX A-X E lt-----gt X-E A X-E B lt-----gt B-X
E
56
Catalysts
Without a catalyst the intermediate converts back
to the reactants and does not proceed forward
(high barrier). Donation of a proton by water or
an acid helps the process move forward. The
active sites of enzymes contain amino acid R
groups, that participate in the catalytic
process as proton donors or proton acceptors.
57
Proton donor/acceptor (Nucleophile/electrophile)
Asp and Glu are negatively charged at pH7.0 and
their side chains are acidic. These side chains
ACCEPT protons which neutralize the charge. Lys,
Arg, His are positively charged at pH 7.0 and
their side chains are basic. These side chains
DONATE protons to neutralize their
charge. Asp/Glu COO- H lt-----------gt COOH
Lys/Arg NH3 lt----------gt NH2 H
Nucleophiles R-OH lt---gt R-O H (hydroxyl) R-SH
lt---gt R-S H (sulphydryl) R-NH3 lt---gt R-NH2
H (amino)
Electrophiles H Proton M Metal ion
Carbonyl
Nucleophiles-groups rich in and capable of
donating electron (attracted to
nucleus) Electrophile- group deficient in
electron (attracted to electron) Reactions are
promoted by proton donors (general acids) or
proton acceptors (general bases). The active
sites of some enzymes contain side groups, that
can participate in the catalytic process as
proton donors or proton acceptors.
58
Acid-base catalysis
Reaction acceleration achieved by catalytic
transfer of proton A general base (B) acts as
proton acceptor to remove proton from OH, NH, CH
(XH) This produces a stronger nucleophilic
reactant (X) A general base(B) removes a proton
from water thereby generating the equivalent of
OH-in neutral solution.
A general acid (BH) can donate a proton. A
covalent bond may break more easily if one of its
atoms is protonated.
59
Acid base catalysis
RNaseA cleavage of RNA
60
xxxxxxx
61
Enzyme Inhibition
62
Enzyme Inhibition
Many molecules inhibit enzymes
63
Competitive Inhibitor
64
Competitive Inhibitor
- Most common
- Inhibitor competes with natural substrate for
binding to active site - Inhibitor similar in structure to natural
substrate and binds active site of enzyme
(reducing effective enzyme conc) - Binds more strongly
- May or may not react
- If reacts, does so very slowly
- Gives info about active site through comparison
of structures
65
Drug targets
66
Gleevec
67
Gleevec How it works
68
HIV protease structure
69
Protease Inhibitors
70
Protease Inhibitor
71
72
Reversible Inhibition (competitive)
- Inhibitor competes with substrates for binding to
active site - Inhibitor is similar in structure to substrate
- binds more strongly
- reacts more slowly
- Increasing I increases EI and reduces E
that is available for substrate binding - Need to constantly keep I high for effective
inhibition (cannot be metabolized away in body) - Slope is larger (multiplied by a)
- Intercept does not change (Vmax is the same)
- KM is larger (multiplied by a)
73
Uncompetitive Inhibitor
Binds only to ES complex but not free enzyme
Binds at location other than active site Does
not look like substrate. Binding of inhibitor
distorts active site thus preventing substrate
binding and catalysis Cannot be competed away by
increasing conc of substrate (Vmax is affected by
I) Increasing I lowers Vmax and lowers Km.
74
- Increasing I
- Lowers Vmax (y-intercept increases)
- Lowers KM (x-intercept decreases)
- Ratio of KM/Vmax is the same (slope)
75
Mixed
- Inhibitor binds E or ES
- Increasing I
- Lowers Vmax (y-intercept increases)
- Raises KM (x-intercept increases)
- Ratio of KM/Vmax is not the same (slope changes)
76
Non-competitive inhibition
Inhibitor binds ES or E It is a special case of
mixed inhibition where Vmax is lowered when I
increases but Km does not change
77
Reversible Inhibition (non-competitive)
A inhibitor binds the enzyme but not in its
active site. It affects the Kcat because
substrate can still bind the active site. Rate of
catalysis is affected
Inh
1/v
Vmax
-Inh
-Inh
1/Vmax (app)
Vmax (app)_
inh
vo
1/2 Vmax
1/Vmax
1/2 Vmax (app)
-1/Km
1/S
Substrate
Km
Km (app)
Vmax is decreased proportional to inhibitor conc
78
(No Transcript)
79
Example
- When a slice of apple is cut, it turns brown-
enzyme o-diphenol oxidase oxidizes phenols in the
apple - Lets determine max rate at which enzyme functions
(Vmax), and Km - 1 When it acts alone (we will use catechol as
substrate. Enz converts this to o-quinone which
is dark and can be measured via absorbance at 540
nm - 2 when it acts in presence of competitive
inhibitor para hydroxy benzoic acid which bind
active site but is not acted upon - when it acts in the presence of a non-competitive
inhibitor- phenylthiourea which binds copper in
the enzyme which is necessary for enzyme activity - Make a supernatant of the apple-enzyme. Measure
color produced (product) - Set up 4 tubes with different conc of cathecol
and a fixed amount of enzyme (apple pulp). - Measure change in absorbance at 1 min intervals
for several minutes and record average change in
absorbance. - Absorbance is directly proportional to product,
we can measure rate of reaction (velocity)
TubeA TubeB TubeC TubeD
S mM mM mM mM
1/S
Vi (DOD)
1/Vi
1/Vmax10 Vmax0.1 -1/Km-0.8 Km1.25mM
80
Example
Each tube also has a fixed amount of PHBA
(competitive inhibitor)
1/Vmax10 Vmax0.1 -1/Km-0.4 Km2.5 mM
TubeA TubeB TubeC TubeD
S mM mM mM mM
1/S
Vi (DOD)
1/Vi
Each tube has a fixed amount of phenylthiourea
(non competitive inhibitor)
TubeA TubeB TubeC TubeD
S mM mM mM mM
1/S
Vi (DOD)
1/Vi
1/Vmax20 Vmax0.05 -1/Km-0.8 Km1.25 mM
81
Irreversible
- Inhibitor
- Binds covalently, or
- Destroys functional group necessary for enzymatic
activity, or - Very stable noncovalent binding
- Suicide Inactivators
- Starts steps of chemical reaction
- Does not make product
- Combines irreversibly with enzyme
82
Regulation of enzymes
- Catalytic activity is increased or decreased
- Enzyme synthesis or degradation
- Covalent modification
- Non-covalent binding and conformational change
(allosteric) - Usually located early in multi-enzyme reaction
pathway - Kinetics differ for allosteric enzymes- sigmoidal
curve and K1/2 instead of Km
- Usually large multiple subunits
- Compare to Hb
- Site for allosteric modulator (R regulatory)
generally different from active site (C
catalytic) - Can be positive or negative
83
xxxxxxx